Figure 1.
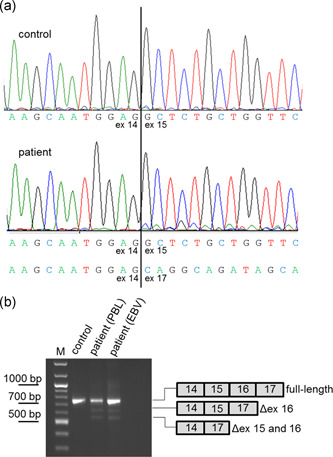
Direct complementary DNA (cDNA) sequencing of patient F8519 carrying NF1 mutation c.1722‐11T>G reveals aberrantly spliced transcripts. (a) Sanger sequence (sense direction) of transcripts isolated from puromycin‐treated short‐term lymphocyte cultures of the patient and a non‐NF1 control individual at the border of exon 14 (ex 14) and exon 15 (ex 15). The wildtype sequence and the sequence deduced from the faint background peaks starting at the beginning of exon 15 in the patient's sequence are given below the electropherograms. This background sequence, which is absent in the control, is derived from exon 17 indicating presence of a small proportion of transcripts lacking exons 15 and 16. (b) RT‐PCR products amplified from transcripts of the patient that were isolated from a puromycin‐treated short‐term lymphocyte culture (PBL) and from EBV‐transformed lymphoblastoid cell line (EBV) also treated with puromycin before cell harvest. Two weak bands coming from a small proportion of transcripts lacking exon 16 only (∆ex 16) or exons 15 and 16 (∆ex 15 and 16) are visible. These bands are absent in the control. To allow for a better separation of the aberrant transcripts from the full‐length transcript, we used in this RT‐PCR experiment instead of our standard primers for direkt cDNA sequencing that generate a product of 1868 bp from the wildtype transcript (Messiaen & Wimmer, 2012) a primer pair that generates a shorter PCR product of the wildtype (693 bp) and aberrant (569 and 491 bp, respectively) transcripts. RT‐PCR, reverse transcriptase polymerase chain reaction
