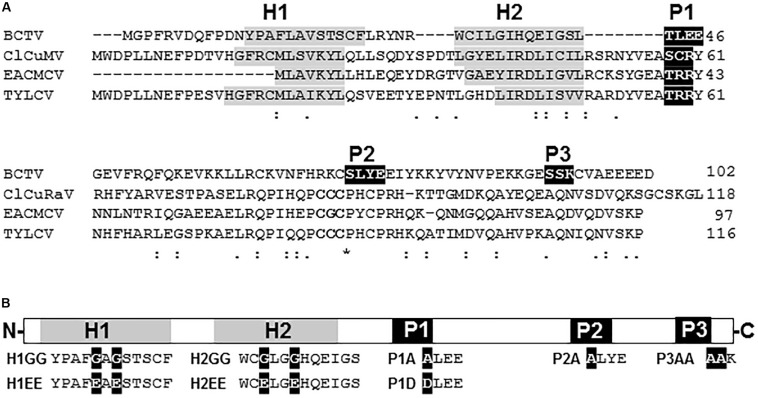
FIGURE 1.
V2 sequence from curtovirus and begomovirus. (A) Alignment of the aminoacid sequences of the V2 proteins from the curtovirus Beet curly top virus (BCTV; M24597) and the begomoviruses, Cotton leaf curl Multan virus (CLCuMV-Fai[PK:Fai2]; AJ496287), East African cassava mosaic Cameroon virus (EACMCV; AF112354) and Tomato yellow leaf curl virus (TYLCV; X15656). Gaps (-) were introduced to optimize the alignment. The positions of the predicted putative phosphorylation motifs P1 (protein kinase CK2/protein kinase C), P2 (protein kinase CK2) and P3 (protein kinase C) are depicted in white letters inside black boxes. The CxC motif from begomoviruses (Padidam et al., 1996; Zrachya et al., 2007) is underlined. The hydrophobic domains (H1 and H2) are shadowed in gray. (B) Schematic view of BCTV V2 aminoacidic sequence. Hydrophobic domains (H1 and H2) are depicted as gray squares. Putative phosphorylation motifs (P1, P2 and P3) are presented as black squares. Amino acid substitutions for each mutant (H1GG, H1EE, H2GG, H2EE, P1A, P1D, P2A, and P3AA) are indicated as white letters in black squares.