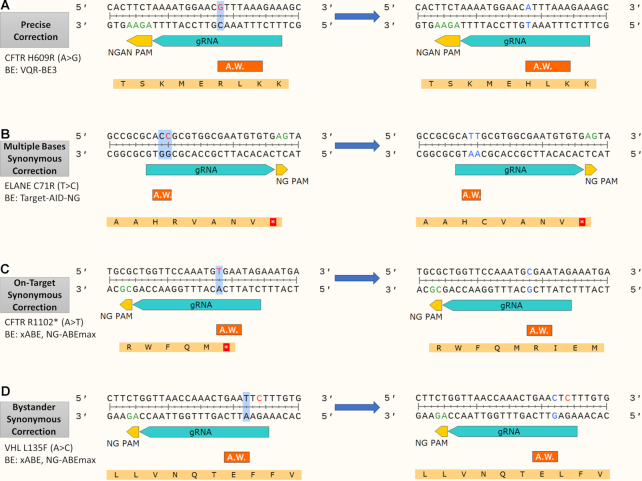
Figure 1.
Base editing correction scenarios. The gRNA and PAM sequences appear in bright blue and yellow, respectively. The major activity window of the base editor is shown as A.W. in orange. The left sequences represent the pathogenic SNV (red) sequences and the right sequences represent the simulated base-edited (blue) sequences. The target nucleotides within the activity window are marked with blue background. (A) Precise correction: a transition mutation precisely repaired by VQR-BE3. (B–D) Synonymous correction scenarios. The resulting DNA sequence does not match the reference allele; however, the translated protein sequence matches. (B) Multiple bases synonymous correction: in addition to the target nucleotide, a bystander nucleotide lies within the activity window and undergoes base editing. (C) On-target synonymous correction: the variant nucleotide (T) is not restored to the reference nucleotide (A), but to another nucleotide (C). The resulted codon, however, is encoded to the reference AA. (D) Bystander synonymous correction: the target nucleotide remains intact while a bystander editing restores the reference protein sequence.