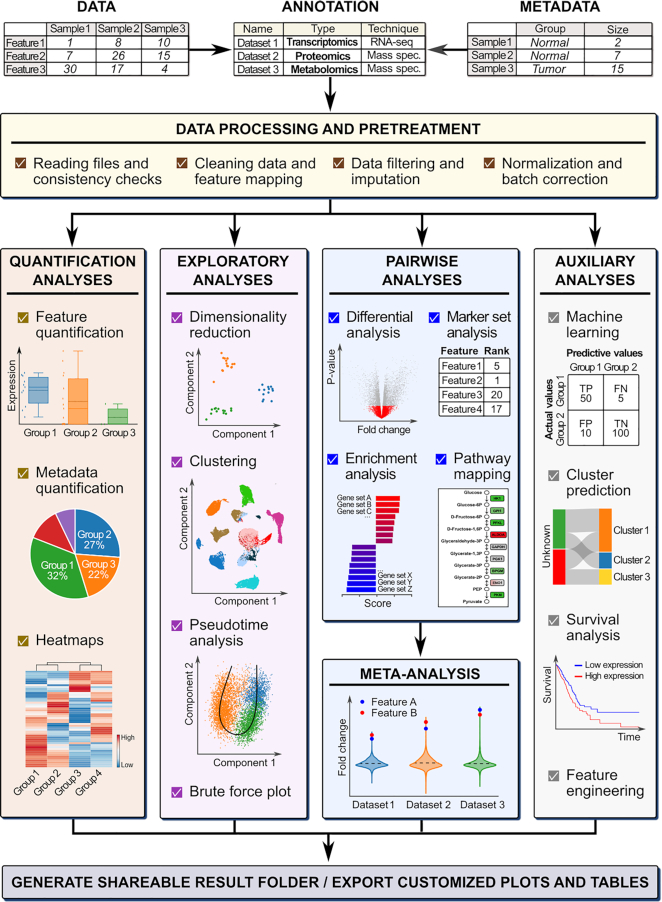
Figure 1.
The BIOMEX workflow. The workflow guides the user through distinct analysis steps. The data (and metadata) need to be uploaded, and each uploaded dataset must be annotated with the relevant information (i.e. the omics data type, technology, feature identifier, etc.). In the processing step, the data is cleaned and consistency checks are performed to verify that the data was uploaded in the correct format. After the feature identifiers are mapped to feature names, the data is filtered and normalized in the pretreatment step. Depending on the omics data type, the data can also be imputed or batch corrected. Once the data is pretreated, it is ready for downstream analysis. The different analyses available in BIOMEX can be divided into five categories: (i) quantification analyses quantify the abundance level of the features in the data (and metadata); (ii) exploratory analyses assist in understanding the underlying structure of the data; (iii) pairwise analyses reveal the functional differences between groups; (iv) meta-analysis combines results from different studies in a singular, unique and robust result and (v) auxiliary analyses (e.g. machine learning and survival analysis). Ultimately, all the analyses can be saved in a self-contained folder that can be shared between scientists and results can be customized and exported either as tables or high quality publication-ready figures. Abbreviations: TP, true positive, FN, false negative, TN, true negative, FP, false positive. Note: The term ‘features’ is used to indicate genes, metabolites and proteins.