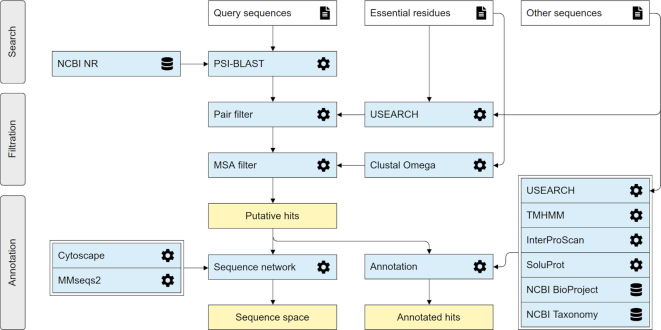
Figure 1.
The EnzymeMiner workflow. The workflow consists of three distinct steps: (i) sequence homology search, (ii) filtration of functional sequences, and (iii) annotation of hits. These steps are executed consecutively and automatically. EnzymeMiner has only two required inputs: (i) query sequences, and (ii) essential residue templates. The Other sequences are optional inputs that allow EnzymeMiner to calculate the sequence identity between these sequences and all the hits. Input files are highlighted by a white background, tools and databases have a light blue background, outputs are highlighted by a yellow background.