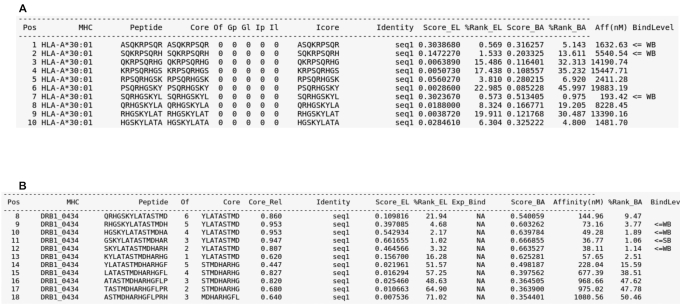
Figure 1.
Example outputs for the NetMHCpan-4.1 and NetMHCIIpan-4.0 tools. (A) Example output for NetMHCpan-4.1, using as input the web server's FASTA sample data and the HLA-A*30:01 allele, with a peptide length of nine and other options set to default. (B) Example output for NetMHCIIpan-4.0, using as input the web server's FASTA sample data and the DRB1*04:34 allele, with all other options set to default. By default, prediction scores are for both methods displayed in terms of a Score_EL (the likelihood of a peptide being an MHC ligand) column and a ‘%Rank_EL’ column (the EL percentile Rank score); if the user selects to include BA predictions, such values are reported as well. The ‘BindLevel’ column displays the presence of Strong Binders (SB) or Weak Binders (WB) amongst the queried peptides. ‘Peptide’ informs the list of peptides that have been interrogated against the selected MHC molecule(s) (exhibited in the ‘MHC’ column). The ‘Pos’ entry refers to the queried peptide's position in the selected FASTA input, and ‘Core’ refers to such peptide's identified binding core. ‘Identity’ is an automatically generated ID that is assigned to the input. Other columns refer to specific properties that depend on the MHC class being employed. For additional details on the interpretation of the different columns of the output, refer to the ‘output format’ page on both web servers homepages.