Figure 1.
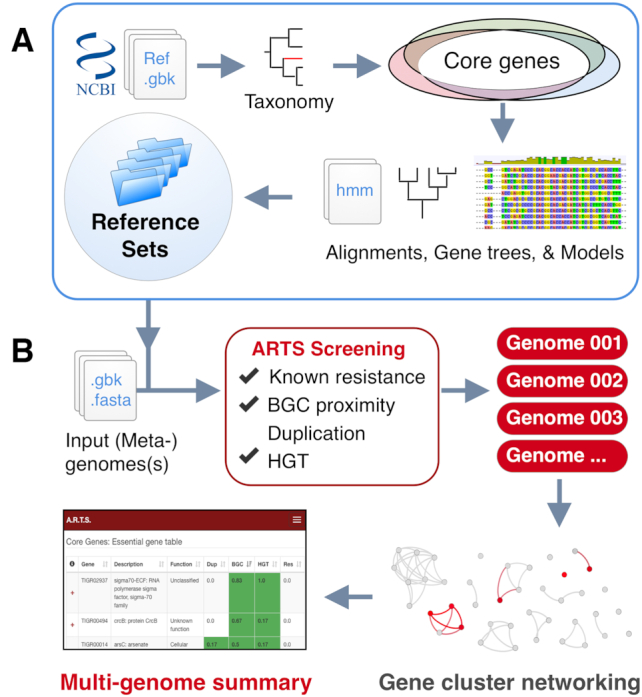
Outline representation of the ARTS pipeline. (A) Basic machinery of creating reference sets. Housekeeping core genes and duplication thresholds are detected per clade of organisms and gene alignments and trees are created for fast HGT detection. (B) Workflow with multi-genome comparative analysis. Input data is screened for ARTS selection criteria. All found BGCs are then subjected to BiG-SCAPE clustering algorithm. Finally, interactive output tables are presented for comparative analysis.
