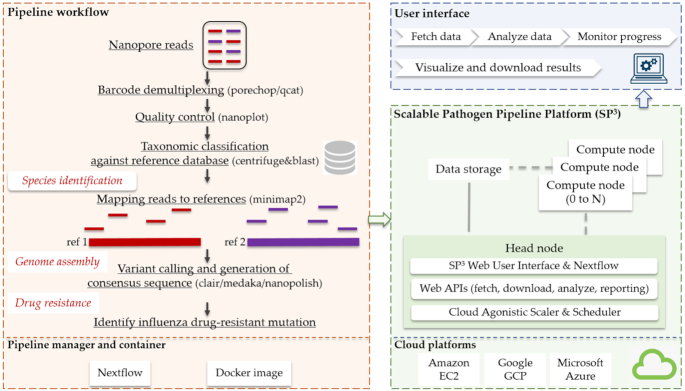
Figure 1.
A schematic overview of NanoSPC. NanoSPC is a scalable, portable, and cloud compatible pipeline that can analyse the Nanopore metagenomic sequencing data. NanoSPC can identify potentially pathogenic viruses and bacteria simultaneously to provide comprehensive characterization of individual samples. The pipeline can also detect single nucleotide variants and assemble high quality complete consensus genome sequences. NanoSPC uses Nextflow pipeline manager and packs all the software dependencies within Docker images (red). NanoSPC can be deployed into the scalable pathogen pipeline platform, enabling elastic computing for high throughput Nanopore data on multiple cloud platforms (green). NanoSPC can be accessed via a web interface to run cloud-based analysis as well as Docker images to analyze data locally (blue).