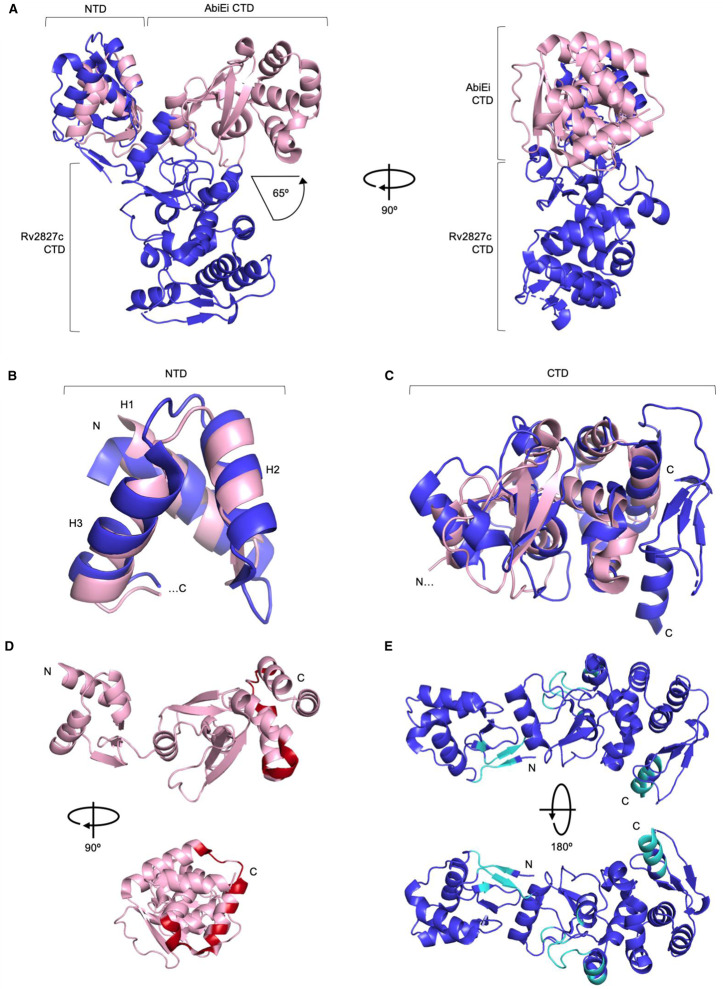
Figure 2. AbiEi and Rv2827c are structurally similar, but have been captured in different positions with differing predicted protein interaction interfaces.
(A) AbiEi (pink) and Rv2827c (blue) in cartoon representation, aligned via the N-terminal winged helix-turn-helix domains, shown as two orthogonal views. The positions of the C-terminal domains diverge at a 65° angle. (B) Close-up structural superposition of the isolated N-terminal helices of AbiEi and Rv2827c, colored as per (A). The three helices (H1–3) of the N-terminal winged helix-turn-helix domains align well. (C) Close-up structural superposition of the isolated C-terminal domains of AbiEi and Rv2827c, colored as per (A). The core secondary structural features of the C-terminal domains approximate to the same positions, but the Rv2827c C-terminal domain has additional features at the C-terminus. (D) AbiEi has C-terminal residues predicted to be involved in making protein–protein interactions, which might allow positive co-operativity in AbiEi monomer binding. AbiEi is in pink cartoon representation with identified interacting residues in red, and is shown in orthogonal views. (E) Rv2827c does not have an equivalent patch of C-terminal interacting residues. Rv2827c is in blue cartoon representation, with identified interacting residues in cyan, and is shown in 180° rotation. Residues were identified using the cons-PPISP server. Rv2827c PDB code: 1ZEL.