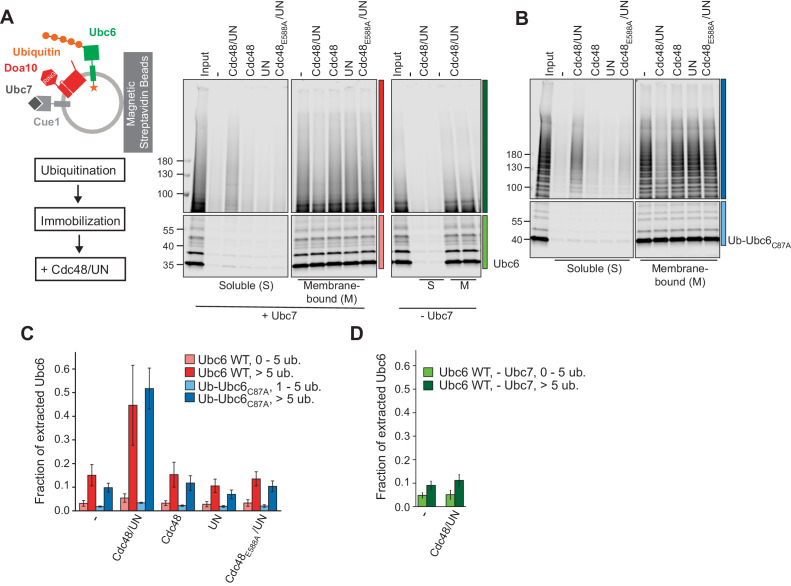
Figure 3. Cdc48-mediated Membrane Extraction of Ubc6.
(A) Extraction of Ubc6 by Cdc48 and Ufd1/Npl4 (UN). After ubiquitination, liposomes were immobilized (Figure 3—figure supplement 1,A to D). One bead equivalent was removed, and bound protein was eluted with SDS sample buffer (Input). Beads were then incubated with the indicated components. Soluble (S) and membrane-bound (M) material were analyzed by SDS-PAGE and fluorescence scanning. Colored bars indicate categorization of ubiquitin chain length as used for quantification in (C) and (D). For better visibility, bottom and top gel parts are scaled differently. See Figure 3—figure supplement 1E for uncut image. Final concentrations: 50 nM Ubc6, 20 nM Doa10, 0.1 µM Cdc48 hexamer, 0.1 µM Ufd1 and Npl4. (B) As in (A), but with Ub-Ubc6C87A instead of Ubc6. See Figure 3—figure supplement 1F for uncut image. (C) Quantification (mean ± SD) of three experiments as in (A) and (B). Ubiquitinated species were categorized according to ubiquitin chain length, as indicated in (A) and (B). The signal in the soluble fraction was normalized to that in the input. (D) Quantification (mean ± SD) of three experiments as in (A), when ubiquitination was performed in the absence of Ubc7.