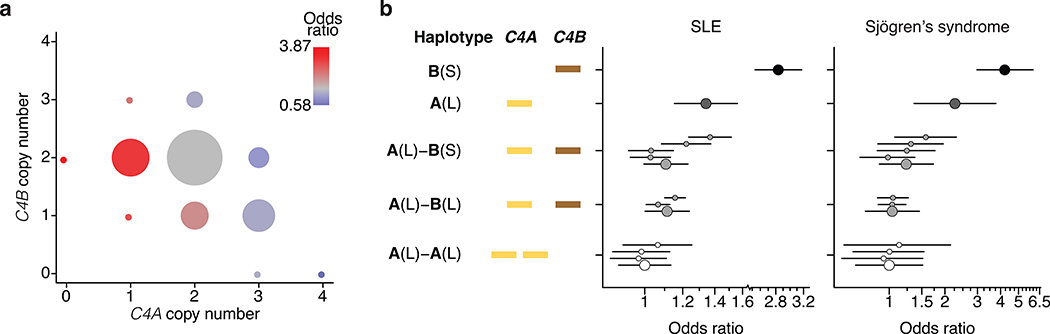
Figure 1. Association of SLE and Sjögren’s syndrome (SjS) with C4 alleles.
(a) Levels of SLE risk associated with 11 common combinations of C4A and C4B gene copy number. The color of each circle reflects the level of SLE risk (odds ratio) associated with a specific combination of C4A and C4B gene copy numbers relative to the most common combination (two copies of C4A and two copies of C4B) in gray. The area of each circle is proportional to the number of individuals with that number of C4A and C4B genes. Paths from left to right on the plot reflect the effect of increasing C4A gene copy number (greatly reduced risk); paths from bottom to top reflect the effect of increasing C4B gene copy number (modestly reduced risk); and diagonal paths from upper left to lower right reflect the effect of exchanging C4B for C4A copies (modestly reduced risk). Data are from analysis of 6,748 SLE cases and 11,516 controls of European ancestry. The odds ratios are reported with confidence intervals in Extended Data Fig. 2c.
(b) SLE and SjS risk associated with common combinations of C4 structural allele and MHC SNP haplotype. For each C4 locus structure, separate odds ratios are reported for each “haplogroup,” i.e., the MHC SNP haplotype background on which the C4 structure segregates. Data are from analyses of 6,748 SLE cases and 11,516 controls for the left plot and 673 SjS cases and 1,153 controls for the right plot. Error bars represent 95% confidence intervals around the effect size estimate for each allele.