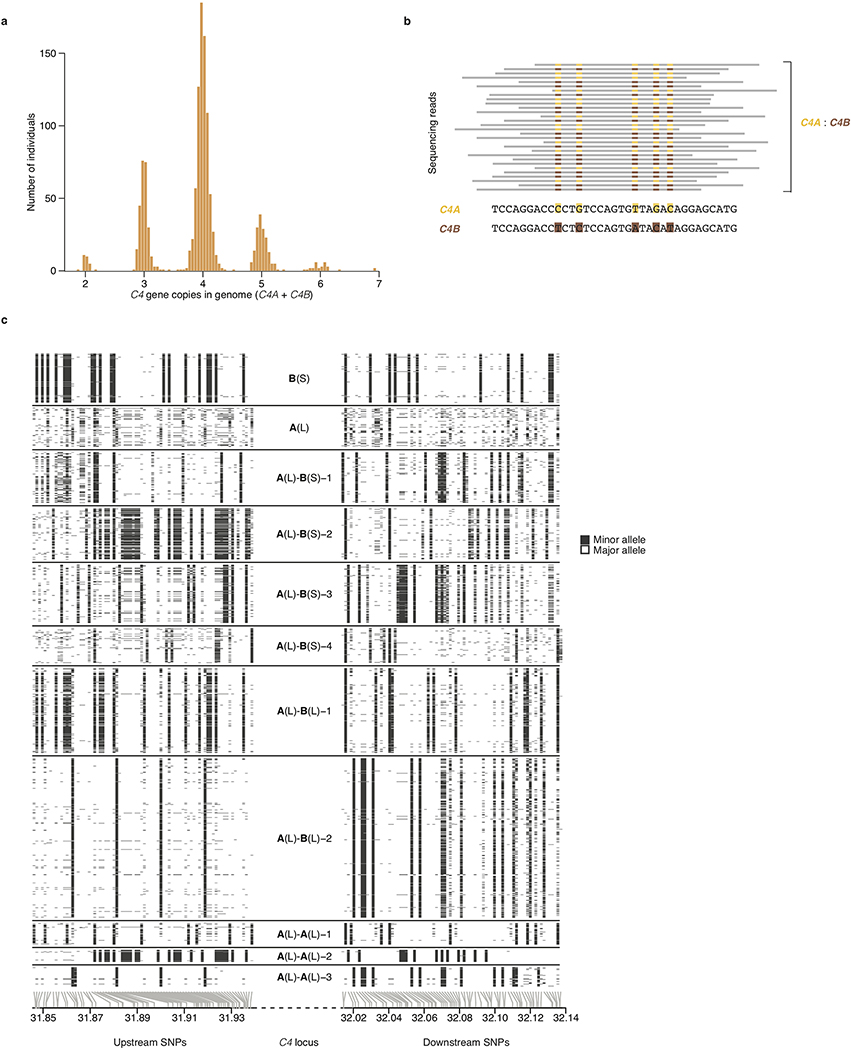
Extended Data Figure 1. A panel of 2,530 reference haplotypes (created from whole-genome sequence data) containing C4 alleles and SNPs across the MHC genomic region enables imputation of C4 alleles into large-scale SNP data.
(a) Distributions (across 1,265 individuals) of total C4 gene copy number (C4A + C4B), as measured from read depth of coverage across the C4 locus, in whole-genome sequencing data.
(b) The relative numbers of reads that overlap sequences specific to C4A or C4B (together with the total C4 gene copy number as in a) are used to infer the underlying copy numbers of the C4A and C4B genes. For example, in an individual with four C4 genes, the presence of equal numbers of reads specific to C4A or C4B suggests the presence of two copies each of C4A and C4B. Precise statistical approaches (including inference of probabilistic dosages), and further approaches for phasing C4 allelic states with nearby SNPs to create reference haplotypes, are described in Methods.
(c) The SNP haplotypes flanking each C4 allele are shown as rows (SNPs as columns), with white and black representing the major and minor allele of each SNP. Gray lines at the bottom indicate the physical location of each SNP along chromosome 6. The differences among the haplotypes are most pronounced closest to C4 (toward the center of the plot), as historical recombination events in the flanking megabases will have caused the haplotypes to be less consistently distinct at greater genomic distances from C4. The patterns indicate that many combinations of C4A and C4B gene copy numbers have arisen recurrently on more than one SNP haplotype, a relationship that can be used in association analyses (Fig. 1b).