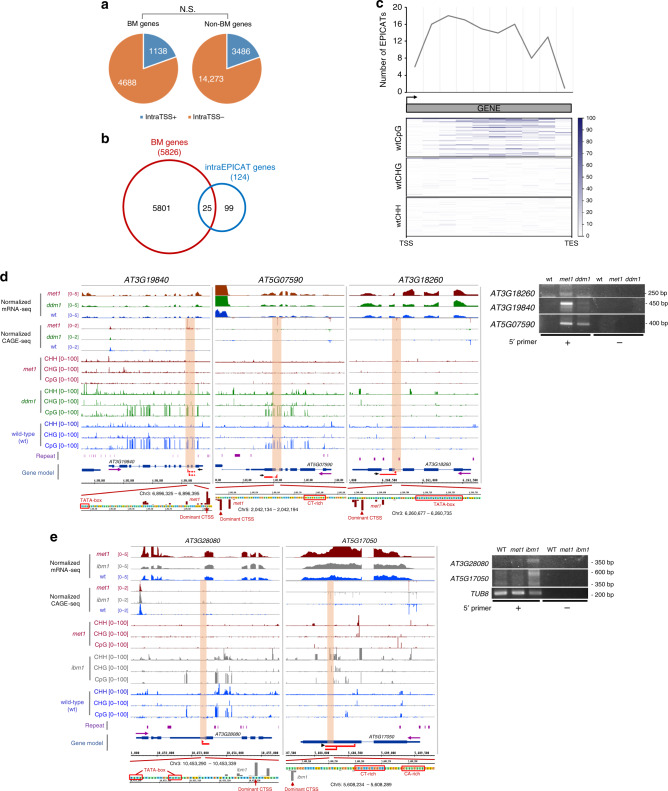
Fig. 4. Gene body methylation (gbM) is not significant for suppressing spurious intragenic transcription inArabidopsis.
a Fractions of body methylated (BM) and non body methylated (Non-BM) genes harboring intragenic TSSs (IntraTSS+) or not (IntraTSS-) in wild-type A. thaliana. N.S.: not significant, Fisher's exact test. b Overlap between body methylated genes (BM genes, red) and genes containing intragenic EPICATs activated in met1 (intraEPICAT genes, blue). c Metaplot showing DNA methylation at genes harboring intragenic EPICATs in met1. Gene body was divided into 10 equal bins. DNA methylation levels (the heatmaps in lower panel) and the number of intragenic EPICATs (upper panel) located within each bin were then calculated. TAIR10-annotated TSS and transcription direction are denoted by black arrow. TSS/TES: Transcription Start/End Sites. d Browser tracks (left) showing the activation of intragenic EPICATs (indicated by orange windows) at AT3G19840, AT5G07590, and AT3G18260, gene loci in the met1 and ddm1 backgrounds. The existence of cryptic transcripts initiated from these EPICATs was confirmed by RACE (right panel). Purple arrows indicate the direction of transcription. Red lines indicate transcripts detected by RACE. Red arrows indicate the end of the transcripts. Black arrows indicate gene specific primers used in RACE. Four independent clones were sequenced for each genotype, and transcript variants detected in at least two clones are shown. Promoter-associated DNA sequences are indicated in red boxes. e Browser tracks (left) showing the activation of intragenic EPICATs (indicated by orange windows) at AT3G28080 and AT5G17050 gene loci in the ibm1 and met1 backgrounds, validated by RACE experiments (right). Data are presented as described in d.