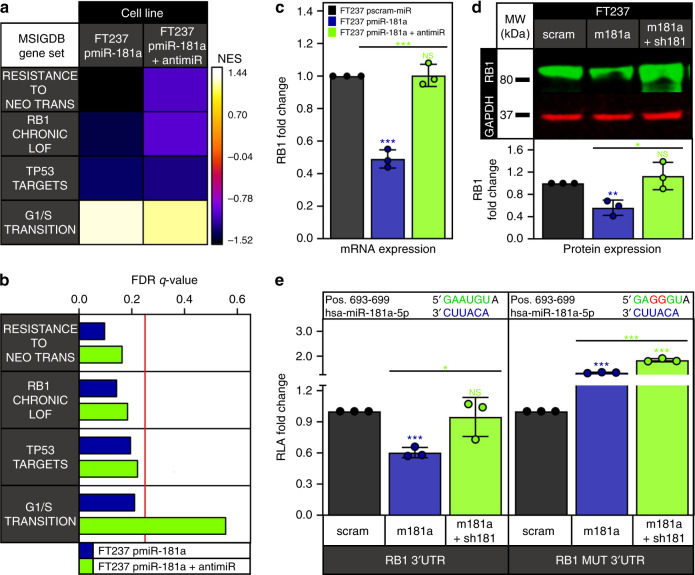
Fig. 6. miR-181a targets RB1 to promote oncogenic transformation of FTSECs.
a Heatmap of normalized enrichment scores for either FT237 pmiR-181a vs pscram-miR or FT237 pmiR-181a + antimiR vs pscram-miR GSEA of significantly altered gene sets associated with RB1 knockdown and oncogenic transformation. b Bar graph of FDR q-values for the FT237 pmiR-181a and pmiR-181a + antimiR GSEA results. The red line indicates the significance cut-off of 0.25. Multiple testing adjustments were made using FDR correction according to default GSEA parameters. c Graph showing RB1 mRNA expression in the FT237 pscram-miR, pmiR-181a, and pmiR-181a + antimiR cells. d Representative western blot of RB1 expression levels in FT237 pscram-miR, pmiR-181a, and pmiR-181a + antimiR cells with quantification below. e Graph showing RB1 3′UTR and mutant 3′UTR relative luciferase activity for the FT237 pscram-miR, pmiR-181a, and pmiR-181a + antimiR cells. miR-181a binding sites within the 3′UTR are depicted in green. Mutated nucleotides within the 3′UTR are depicted in red. All data are representative of N = 3 independent experiments unless otherwise stated. The measure of center for the error bars is given as the mean value unless otherwise stated. The statistical test used for data analysis is the two-sided Student’s t test unless otherwise stated. Error bars indicate ± standard deviation unless otherwise stated. *p < 0.05, **p < 0.005, ***p < 0.0005. Full western blots are shown in Supplementary Fig. 11.