Fig. 4.
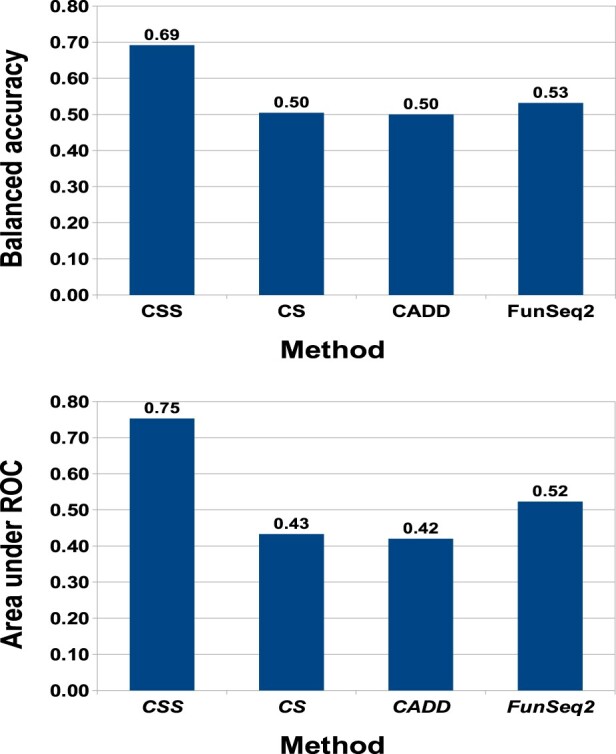
Comparison between CScape-somatic performance in LOCO-CV (non-coding regions, COSMIC data) with prediction results for CScape, CADD and FunSeq2 on the same examples (CSS= CScape-somatic and CS= CScape). Top: CScape-somatic dramatically outperforms other methods on the COSMIC training data with accuracy over 69%. Of the other methods, only FunSeq2 appears to yield prediction accuracy better than chance, at 52.7%. The remaining methods fare poorly, including the original CScape. Bottom: We see the same trend with ROC scores, as CScape-somatic yields satisfactory ranking performance of 0.75, whereas only FunSeq2 yields rankings better than chance
