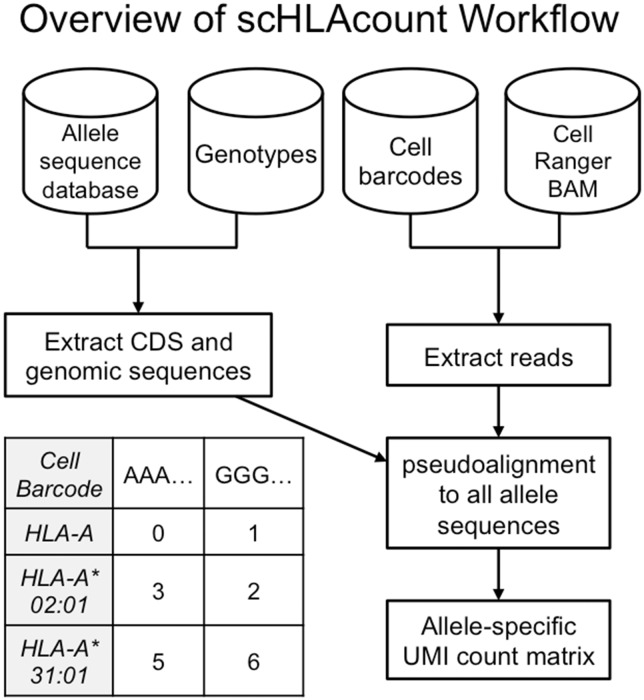
Fig. 1.
scHLAcount takes as input an allele sequence database (e.g. IMGT/HLA), genotypes for the sample being evaluated, cell barcodes and aligned reads (e.g. BAM file from Cell Ranger). Allele sequences and relevant reads are extracted, and pseudoalignment is used to produce an allele-specific molecule count matrix. A snippet of the output matrix is shown for two cell barcodes and one gene (HLA-A) with two alleles