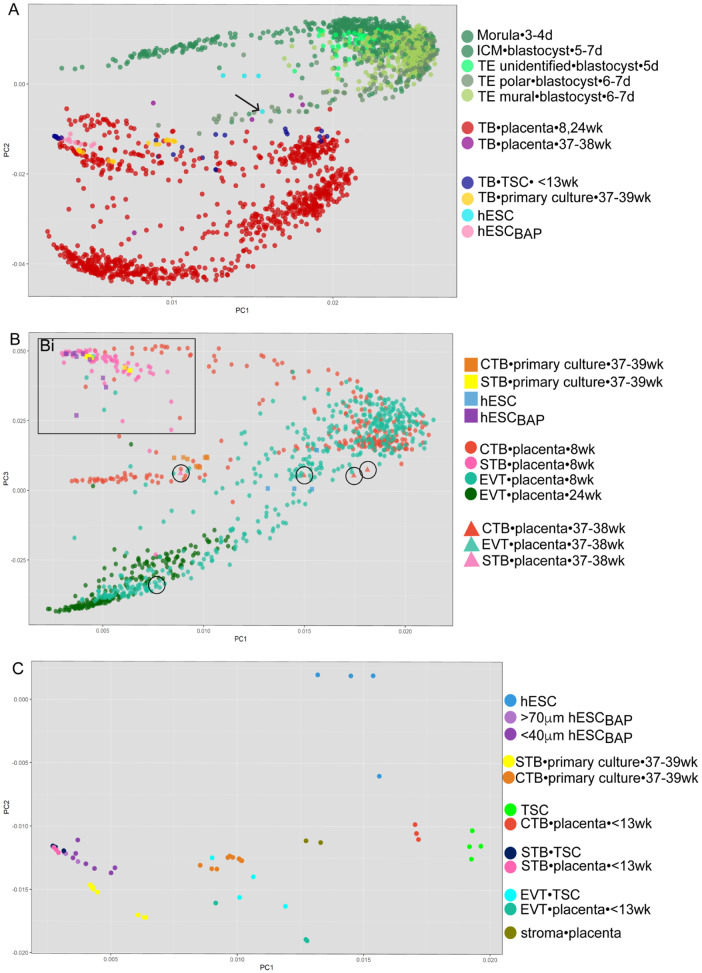
Figure 2.
Principal component analysis comparing five human trophoblast RNAseq datasets from the following sources: morula-blastocyst stage human embryos (Petropoulos et al., 2016a), trophoblast stem cells (Okae et al., 2018), placentas (Pavlicev et al., 2017; Liu et al., 2018), and embryonic stem cells and primary trophoblast cultures (Yabe et al., 2016). For each datapoint, cell type•cell source•gestational age are indicated in the legend on the right. (A) Principal component (PC) 1 and 2 separated by cell origin. hESCBAP cluster separately from cells of the blastocyst and from term trophoblast. Arrow indicates an hESC sample with morphological signs of spontaneous differentiation. (B) hESCBAP and trophoblast isolated from first trimester placentas do not cluster with primary trophoblast lineages from term trophoblast isolated from term placenta (circles). The hESCBAP cluster closer to 8 weeks CTB and STB than to 8 weeks or 24 weeks EVT. (C) STB derived from TSCs, hESCBAP, and first trimester STB cluster together. First trimester CTB and TSCs cluster close to one another yet distinctly from hESCBAP. The hESCBAP cluster closely with STB from the first trimester across multiple datasets. CTB, cytotrophoblast; D, days since conception; EPI, epiblast; EVT, extravillous trophoblast; hESC, human embryonic stem cell; embryonic stem cell; hESCBAP, hESC differentiated to TB with BAP; ICM, inner cell mass; PE, primitive endoderm; STB, syncytiotrophoblast; TB, trophoblast; TE, trophectoderm; TSC, trophoblast stem cell; Wk, weeks since last menstrual period.