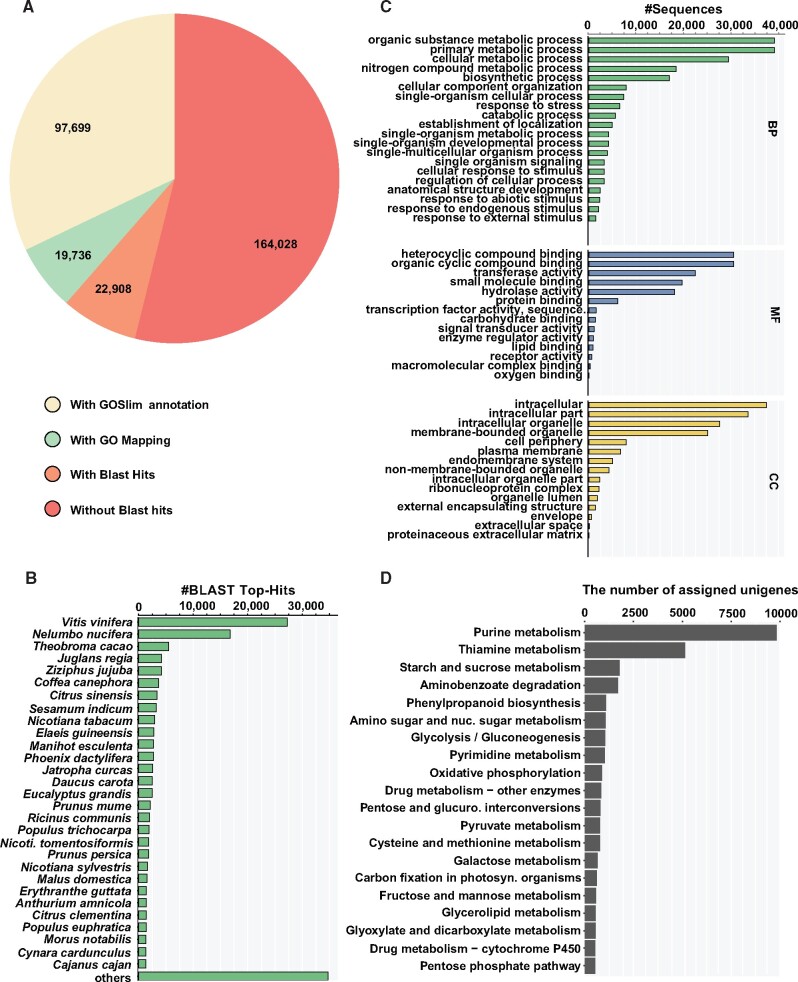
Figure 4.
Functional annotation of the de novo transcriptome assembly of Cornus officinalis. (A) Blastx-based data distribution summary for the annotated transcripts of C. officinalis. (B) Blastx top-hit species distribution for the assembled transcripts. (C) Blastx-based GO terms for all the annotated transcripts are summarized in three broad categories; biological processes (BP), molecular function (MF), and cellular components (CC); and (D) top 20 KEGG pathways represented by the annotated transcripts of C. officinalis based on the number of assigned transcripts.