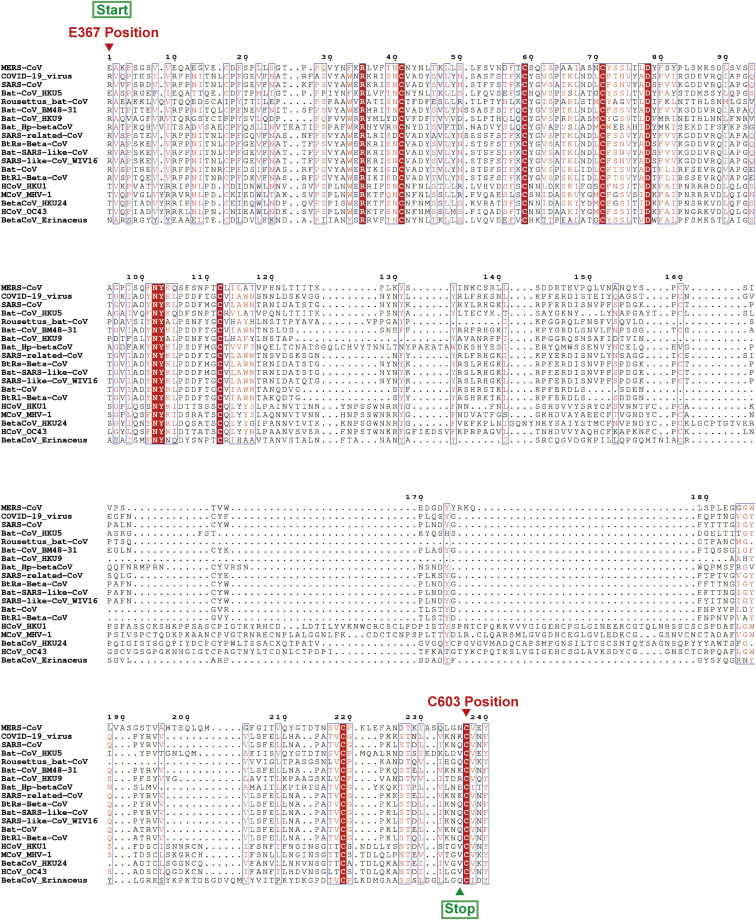
Figure S2.
Amino Acid Alignment of 19 RBDs from Beta-CoV Genera, Related to Figures 3 and 4
The sequences are aligned based on a MUSCLE alignment. Conserved residues are highlighted in red. To construct the RBD-sc-dimer, the start and stop residues are highlighted by text in green boxes with arrow mark. The highly conserved C603 position of MERS-CoV S protein is highlighted by text in red with arrow mark. The sequence alignment was generated with Clustal X and ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/index.php) (Robert and Gouet, 2014).