Abstract
The interferon (IFN)-mediated activation of the Janus kinase (JAK)-signal transducer and activator of transcription 1 (STAT1) signaling is crucial for cell sensitivity to ionizing radiation. Several preclinical studies have reported that the IFN/STAT1 pathway mediates radioresistance in the tumor microenvironment by shielding the immune responses and activating survival signaling pathways. This review focuses on the oncogenic function of the IFN/STAT1 pathway, emphasizing the major signaling pathway in radiation sensitization. Furthermore, it highlights the possibility of mediatory roles of the IFN/STAT1 pathway as a prognostic therapeutic target in the modulation of resistance to radiotherapy and chemotherapy. MicroRNA involved in the regulation of the IFN/STAT1 pathway is also discussed. A better understanding of radiation-induced IFN/STAT1 signaling will open new opportunities for the development of novel therapeutic strategies, as well as define new approaches to enhance radio-immunotherapy efficacy in the treatment of various types of cancers.
Keywords: interferon-gamma signaling, signal transducer and activator of transcription 1, tumor radioresistance, IFN/STAT1, microRNA
Introduction
Radiation therapy (RT) is a widely applied modern technique in cancer therapy, which directly induces DNA damage and indirectly removes cancer cells by inducing cytotoxic pathways.1 Recently, the preclinical data suggest that the generation of anti-tumor immune response may also play an important role in the effectiveness of RT.2 RT combined with immunotherapy shows a synergic potentiation to enhance systemic antitumor immunity.3 However, the radioresistance of tumor cells, as well as the specific radioprotection of normal tissues, are the main challenges of RT and need to be addressed in treatments aimed at preventing tumor recurrence. A recent study reported intrinsic tumor cell radioresistance, genotoxic stress, and tumor microenvironment (TME) factors as the main obstructions of effective RT.4
Interferons (IFNs) are a group of pleiotropic cytokines in the tumor-suppressive network which have been extensively studied in the context of viral infections, anti-tumor immune responses,5 and RT alone or with immunotherapy.6 As the master transcription factor of IFN-related intracellular signaling, signal transducer and activator of transcriptions (STATs), has been widely explored in cancer progression. STAT1 and STAT2 are the first STAT proteins identified in the IFN signal transduction pathways that have an essential role in suppressing tumor cell proliferation, differentiation, apoptosis and angiogenesis. Similarly, in the STATs family, STAT3 and STAT5 are the important players in the cancer development, whereas STAT1 is predominantly viewed as a tumor suppressor.7,8 However, the bulk of evidence indicated that aberrant STAT1 activation triggers radioresistant properties in several cancers, including human head and neck squamous cell carcinoma,9 myeloma,10 renal cell carcinoma,11 and breast cancer.12 Moreover, the suppression of STAT1 expression reversed tumor radioresistance and malignant progression in renal cell carcinoma11 and nasopharyngeal cancer.13 In comparison with other STAT isoforms, STAT1 is the main transcription factor that acts as a critical control point of the phenomenon of therapeutic resistance to ionizing radiation (IR) in different types of cancer cells. Thus, understanding the mechanisms of the interaction between the IFN/STAT1 pathway and cancer cells could lead to the development of highly tailored radio-immunotherapy.
In this review, we summarized the current knowledge of radioresistance mediated by the IFN/STAT1 pathway, with a particular focus on the oncogenic function of STAT1 signaling and its interaction with tumor cells in the TME. Furthermore, this review aimed to explore the possible roles of the IFN/STAT1 pathway as a prognostic therapeutic target in radioresistance.
The Canonical IFN/STAT1 Signaling Pathway in Tumor Radioresistance
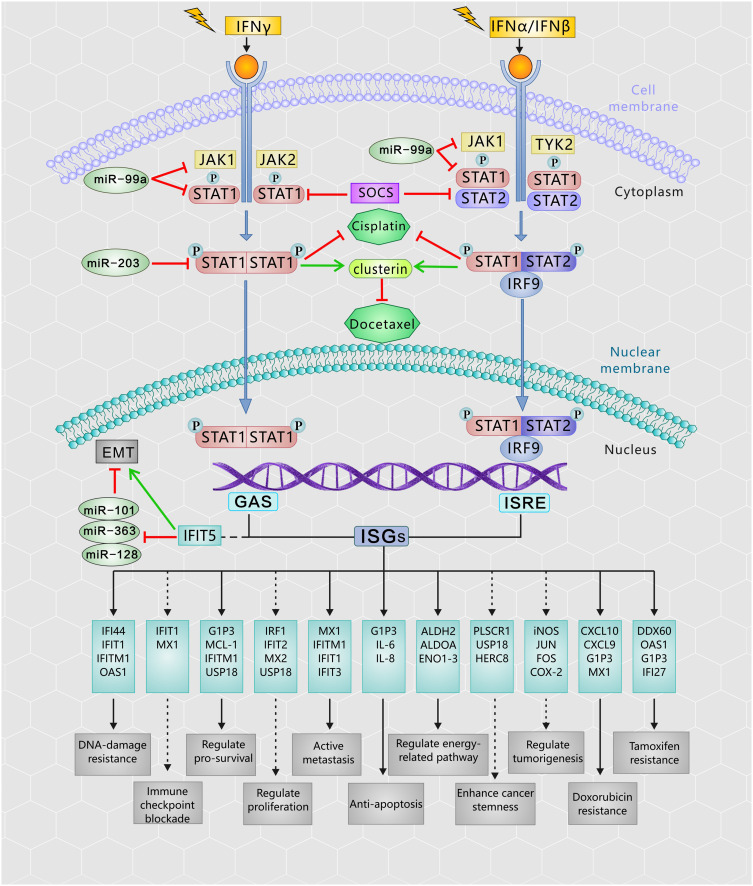
IFN/STAT1 pathway-induced inherent and/or acquired resistance to the cytotoxic effects of RT might impede the effectiveness of cancer treatment. A literature review showed that the IFN/STAT1 pathway in the TME has different functions and regulates several proto-oncogenes, such as inducible nitric oxide synthase (iNOS), JUN, FOS, and cyclooxygenase 2 (COX-2).7,14,15 RT has been proposed to enhance innate and adaptive antitumor immunity, as well as increase the expression levels of IFNs in the TME. Figure 1 presents the RT-activated IFN/Janus kinase (JAK)/STAT1 pathway in detail. Specifically, Type I IFN (IFN-α and IFN-β) binds to interferon Type I receptors (IFNAR1 and IFNAR2), thereby activating the downstream signaling components, JAK1 and tyrosine kinase 2. These kinases phosphorylate STAT1 and STAT2 in the cytoplasm and assemble a heterotrimeric transcription factor complex known as the interferon-stimulated gene factor 3 (ISGF3) protein complexes. ISGF3 is an essential mediator of IFN signaling comprised interferon regulatory factor 9, STAT1, STAT2, and STAT3.16 ISGF3 translocates into the nucleus to bind interferon-stimulated response elements and activate the expression of interferon-stimulated genes (ISGs) transcription.17,18 Type II IFN (IFN-γ) binds to a specific receptor, the interferon Type II receptor (IFNGR), and then participates in phosphorylation-dependent activation of JAK1 and JAK2. The phosphorylated form of STAT1 is a homodimer able to translocate into the nucleus, where it activates the transcription of ISGs by binding to the interferon-γ-activated sequence.17,19 As a result of Type I/II IFN signaling, ISGs represent a diverse group of genes involved in many cellular functions, such as transcription, apoptosis, and cell cycle regulation.17
Figure 1.
The RT activates the IFNs-JAK/STAT1 pathway in cancer cells. After exposure to the IR, Type I and II IFN bind to their respective receptors to induce a number of signal transduction cascades, then the JAK kinase and the STAT1 being recruited to the receptor and phosphorylated in cytoplasm. The phosphorylated STAT1 triggers the heterodimers (ISGF3) or homodimers, and migrate to the nucleus. These complex bind to ISRE or interferon-gamma-activated sequence (GAS) gene promoter elements, respectively drives the downstream IFN-regulated genes expression and perform cellular functions, which are associated with radioresistance. Phosphorylated STAT1 also contributes to develop cisplatin resistance, as well as upregulate clustering in docetaxel-resistant cells. The miRNAs and SOCS proteins functions as the major regulators to control of the IFNs/STAT1 pathway.
Abbreviations: EMT, epithelial-to-mesenchymal transition; GAS, interferon-gamma-activated sequence; IR, ionizing radiation; IRF9, interferon regulatory factor 9; ISGF3, interferon-stimulated gene factor 3; ISRE, interferon-stimulated response element; IFN, interferon; JAK, Janus kinase; miRNA, microRNA; RT, radiation therapy; SOCS, suppressor of cytokine signaling; STAT1, signal transducer and activation of transcription 1.
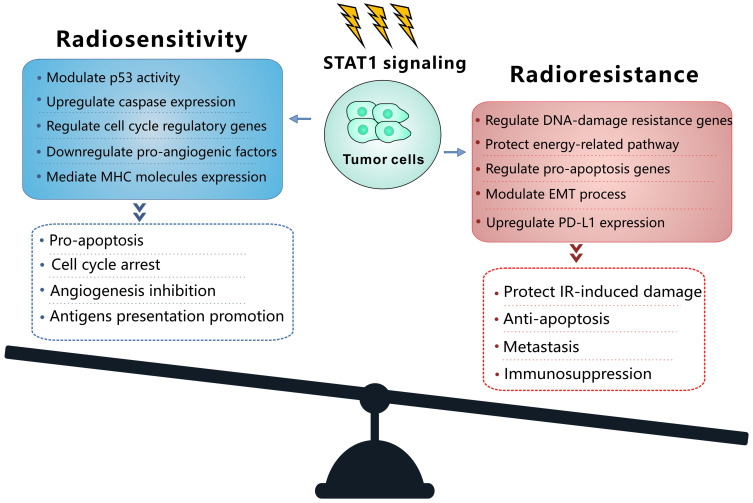
IFNs coordinate tumor-immune system interactions by activating the downstream JAK/STAT1 pathway.7 Any abrogation of Type I IFN signaling in cancer cells enhanced tumor response to IR.20,21 In antigen-presenting cells, IFNs upregulate the major histocompatibility complex class I molecules in STAT1-dependent manner, which effectively promotes the presentation of tumor antigens, and is therefore essential for generating adaptive immune responses.20 Besides, the IFN-γ secreted by T-cells can upregulate the expression of some angiogenic chemokine in tumor-associated macrophages (TAMs), such as CXCL9, CXCL10, and CXCL11.22 However, a wide array of these ISGs is now considered to be tumor promoters and is used to determine the biological consequence of the IFN/STAT1 pathway. Emerging data indicate that Type I IFN can protect cancer cells from CD8+ T-cell-mediated toxicity after RT by upregulation of the immune checkpoints and downregulation of anti-apoptotic and MHC class I molecules.23,24 Recently, serpin family B member 9 (SERPINB9) transcription factor is introduced as a key mediator of enhanced susceptibility to T cell-mediated killing after radiation.21 Activation of Type I IFN signaling and induction of SERPINB9 protects cancer cells from CD8+ T cell-mediated cytotoxicity in a dose-dependent fashion indicating that IFN signaling has a paradoxical effect on tumor control after RT.21 In the TME of radioresistant cells, most STAT1 is phosphorylated and initiates multiple gene expression. These genes mediate diverse cellular functions, such as immune modulation, the regulation of cell proliferation, and cell apoptosis control. Interestingly, STAT1 has a dual function in the TME of radioresistant cells; it can promote the phenotype of radioresistant cancer cells and tumor metastasis. Previous study indicated that STAT1 downregulates the expression of the p53 inhibitor murine double minute 2 (MDM2) oncogene and acts as p53 co-activator.25 MDM2 is the negative regulator of p53 tumor suppressor gene, that p53-MDM2 pathway represents well-known targets for RT.26 Contrarily, it can also promote apoptosis of cancer cells.27 The “Friend” or “Foe” function of STAT1 signaling in the IFN/STAT1 pathway is illustrated in Figure 2. The STAT1 signaling induced by IR does not play a “Foe” role. Thus, STAT1 may directly participate in DNA damage repair.
Figure 2.
Model of balance between the mechanism and the cellular function of the STAT1 signaling to promote radiosensitivity and radioresistance. In the radioresistant cancer cells, IR activates STAT1 signaling and trigger the function of radioresistance or radiosensitivity.
Abbreviations: EMT, epithelial-to-mesenchymal transition; IR, ionizing radiation; MHC, major histocompatibility complex; PD-L1, programmed death ligand-1; STAT1, signal transducer and activation of transcription 1.
Extrinsic Determinants in Tumor Radioresistance
Regulation of Immunosuppressive Factors
Recent studies have highlighted that sustained IFNs expression may induce many inhibitory factors that stimulate tumor growth in the STAT1-dependent pathway, especially by triggering programmed death-ligand 1 (PD-L1).28,29 PD-L1 is a co-signaling molecule expressed on tumor cells or other infiltrating immune cells, which can initiate T-cells apoptosis with the engagement of programmed death 1 expressed on cytotoxic T-lymphocytes.30 PD-L1 expressed in several human hematopoietic tumor cell lines also confers resistance to natural killer cell-mediated lysis. In IFN-γ-induced JAK/STAT1 signaling pathway, the silencing of JAK1 and JAK2 was shown to reverse the resistant phenotype of tumor cells.31 Thus, these modifications in the TME balance of tumor cells can rapidly promote resistance to RT. It is clear that combination therapy of RT with anti-PD-L1 is one of the modern treatment methods to overcome resistance to checkpoint inhibition in RT, which may lead to the downregulation of genes involved in antitumor responses, T-cell exhaustion, antigen presentation, and chemokine production in TME.21,32 For example, the upregulation of PD-L1 in stage II melanoma was determined to be a mechanism involved in the resistance to RT.32 In a PD-L1-dependent manner, this resistance programming can be orchestrated by Type II IFN/STAT1 signaling due to the activation of ISGs, as interferon-induced protein with tetratricopeptide repeats (IFITs) and interferon-induced GTP-binding protein (MX1) families.32 IFNGR and IFNAR knockout experiments showed that IFNs were basic for melanoma cells to maintain both PD-L1-dependent and PD-L1-independent resistance.32 In both tumor cells and stromal cells, IFN/STAT1 pathway stimulated the indoleamine-2,3-dioxygenase (IDO), resulting in progressive immune dysfunction and tumorigenesis. In TME, elevated IDO activity may lead to an impaired T-cells activation. For example, human dendritic cells can mediate IDO-dependent inhibition of T-cells proliferation in the model of mixed leukocyte reaction.33 Moreover, in vitro study demonstrated that IDO-transfected fibrosarcoma cells suppressed T-cell proliferation.34 Blocking of IDO expression may improve the effectiveness of antitumor therapy, such as RT.35 Therefore, the results of these studies clearly show that the IFN/STAT1 pathway promotes the development of radioresistance in cancer cells by inducing immunosuppressive factors in the TME.
Regulation of Immunosuppressive Cells
The presence of radioresistant immunosuppressive cells in TME such as myeloid-derived suppressor cells (MDSCs), regulatory T-cells (Tregs), and TAMs with the M2 phenotype are also the reduced the efficiency of the effective RT.4,36 Through suppressing the adaptive T-cell-mediated antitumor immunity, these cells can promote tumors to immune tolerance.37 Multiple studies have noted that IFN/STAT1 pathway plays a role in the recruitment and activation of immune inhibitory cells. In this view, IFNs are critical for the formation of a tumor radioresistant environment. The MDSCs in breast cancer are a well-evaluated example of Type II IFN/STAT1 pathway’s role that active the immunosuppressive cells and enhance the suppressive function of MDSCs by inducing the expression of pro-inflammatory tumor necrosis factor α, transforming growth factor β, and interleukin-13, STAT1 signaling can recruit the MDSCs in inflamed TME.38 Furthermore, Type II IFN was shown to induce a higher level of iNOS, arginase I, NO production, and arginase activity in both MDSCs and the TAM, which are a response to immune inhibition and T-cell deletion in several cancer models.39 Similarly, in vivo study shows that Tregs with STAT1 deficiency failed to control allograft rejection.40 In a mice model, the absence of certain STAT1 transcription factors reduced the frequency of MC38 tumor-associated interleukin (IL)-10+ Tregs in the colon carcinoma cells.41 Furthermore, IDO expression in melanoma cells enhanced the recruitment of Tregs to evade immune recognition.42 Overall, the ability of the IFN/STAT1 pathway to regulate T-cell function displays a high environmental dependence and variation among tumor types and factors. However, the study findings presented indicate that the IFN/STAT1 pathway is a potential target to improve the effect of cancer RT by recruiting immune inhibitory cells and inhibiting effector T-cells. The regulation of Tregs by the IFN/STAT1 pathway is controversial, however, and more research is needed to elucidate the role of Tregs function in the TME of radioresistant cells.
Enhancement of Radioresistance in Cancer Stem Cells
Cancer stem cells (CSCs), with strong self-renewal capability, have been demonstrated to contribute to tumorigenesis, therapy resistance, and also metastasis, which could result in RT failure.43 Recently, studies have indicated a novel function of STAT1 associated with CSCs. The upregulation of active STAT1 and many ISGs, such as PLSCR1, USP18, and HERC8, is involved in the resistance of CSCs to RT.44,45 A detailed overview of the ISGs associated with tumor radioresistance in the IFN/STAT1 pathway is illustrated in Figure 1 and Table 1. These ISGs, together with STAT1, activate the canonical Type I IFN signaling pathway in cooperation with CD95 in the stemness of CSCs.44 The knockdown of STAT1 is considered one of the best treatment methods for radioresistant cancer cells, which is achieved by reducing the non-apoptotic activities of CD95.44 In addition, higher apoptosis rates were observed in irradiated CD24−/low/CD44+ breast CSCs that treated with STAT1 inhibitor. These cells show that STAT1 inhibited irradiation-induced apoptosis in cancer-initiating cells, resulting in resistance of the breast cancer cells to RT.45 After 4 Gy irradiation, the radioresistant glioma stem cells expressed the highly unregulated STAT1 with some DNA metabolism-related genes.46 Based on these findings, one potential link between STAT1 and radioresistance is presumably through the increased stemness of CSCs, which is mediated by STAT1-activated genes. Targeting the Type I IFN/STAT1 pathway in CSCs may have a broad therapeutic application to rescue cancer radioresistance.
Table 1.
STAT1-Activated Genes in Tumor Radioresistance
| Name | Radiotherapy | Total Dose (Gy) | Function | Drug Resistance | Cancer | References |
|---|---|---|---|---|---|---|
| IFNStimulated Genes | ||||||
| CXCL10 | γ-RAD | 4 or 8 | Antigen presentation | DOXO | M | [10] |
| IFI27 | γ-RAD | 4 or 8 | Apoptosis regulation | DOXO | M | [10] |
| X-ray | 64 | TCT | BC | [67] | ||
| IFI44 | X-ray | 40 | Proliferative regulation | DOXO | SCC | [12] |
| IFI44L | γ-RAD | 4 or 8 | Tumor cell survival promotion | DOXO | M | [10] |
| IFI6 | γ-RAD | 4 or 8 | Apoptosis regulation | DOXO | M | [10] |
| X-ray | 64 | TCT | BC | [67] | ||
| IFIT1 | γ-RAD | 4 or 8 | Anti-viral response | DOXO | M | [10] |
| X-ray | 20 | SCC | [82] | |||
| X-ray | 20 | MM | [83] | |||
| IFIT3 | X-ray | 20 | Anti-viral response | DOXO | SCC | [82] |
| MM | [83] | |||||
| IFITM1 | X-ray | 40 | Anti-viral response | DOXO | SCC | [12] |
| 20 | SCC | [82] | ||||
| 20 | MM | [83] | ||||
| IRF7 | γ-RAD | 4 or 8 | Transcriptional activation | DOXO | M | [10] |
| ISG15 | X-ray | 40 | Protein degradation | DOXO | SCC | [12] |
| MCL-1 | X-ray | 40 | Apoptosis regulation | DOXO | SCC | [12] |
| MX1 | γ-RAD | 4 or 8 | Anti-viral response | DOXO | M | [10] |
| X-ray | 40 | SCC | [12] | |||
| 20 | SCC | [82] | ||||
| 20 | MM | [83] | ||||
| OAS1 | X-ray | 64 | Anti-viral response | TCT | BC | [67] |
| OASL | X-ray | 20 | Anti-viral response | DOXO | SCC | [82] |
| USP18 | X-ray | 40 | Protein modification | DOXO | SCC | [12] |
| Tumor-Associated Genes | ||||||
| JUN | γ-RAD | 64 | Proliferation regulation | – | NPC | [84] |
| FOS | γ-RAD | 64 | Proliferation regulation | – | NPC | [84] |
| Glycolysis Pathway-Related Genes | ||||||
| ALDH1A1 | X-ray | 30 | Oxidation of HAc to acetate | – | SCC | [62] |
| ALDH2 | X-ray | 30 | Oxidation of HAc to acetate | – | SCC | [62] |
| ALDOA | X-ray | 30 | Oxidation of GAPD | – | SCC | [62] |
| ENO1 | X-ray | 30 | Dehydration of PCK | – | SCC | [62] |
| ENO2 | X-ray | 30 | Dehydration o of PCK | – | SCC | [62] |
| ENO3 | X-ray | 30 | Dehydration of PCK | – | SCC | [62] |
| GAPDH | X-ray | 30 | Oxidation of GAPDH | – | SCC | [62] |
| GPI | X-ray | 30 | Conversion of GPI | – | SCC | [62] |
| LDHA | X-ray | 30 | Reduction of pyr to lactate | – | SCC | [62] |
| LDHAL6B | X-ray | 30 | Reduction of pyr to lactate | – | SCC | [62] |
| LDHB | X-ray | 30 | Reduction of pyr to lactate | – | SCC | [62] |
| PGAM1 | X-ray | 30 | Phosphoryl transferring | – | SCC | [62] |
| PGK1 | X-ray | 30 | Phosphoryl transferring | – | SCC | [62] |
| PGM1 | X-ray | 30 | Interconversion of G1P and G6PD | – | SCC | [62] |
| PKM2 | X-ray | 30 | Phosphoryl transferring | – | SCC | [62] |
| TPI1 | X-ray | 30 | Interconversion of DHAP | – | SCC | [62] |
| Oxidative Phosphorylation-Related Genes | ||||||
| ATP5A1 | X-ray | 30 | Synthesis of ATP from ADP | – | SCC | [62] |
| ATP5O | X-ray | 30 | Synthesis of ATP from ADP | – | SCC | [62] |
| ATP5B | X-ray | 30 | Synthesis of ATP from ADP | – | SCC | [62] |
| COX4I1 | X-ray | 30 | Oxidation of cyc | – | SCC | [62] |
| SDHA | X-ray | 30 | Oxidation of NADH | – | SCC | [62] |
| MDH2 | X-ray | 30 | Oxidation of MDH | – | SCC | [62] |
Abbreviations: ALDH1A1, aldehyde dehydrogenase 1 family member A1; ALDH2, aldehyde dehydrogenase 2; ALDOA, fructose-bisphosphate aldolase A; ATP5A1, ATP synthase, H+ transporting, mitochondrial F1complex, alpha subunit 1, cardiac muscle; ATP5PO, ATP Synthase Peripheral Stalk Subunit OSCP; ATP5B, ATP synthase subunit beta; BC, breast cancer; COX4I1, cytochrome c oxidase subunit IV isoform 1; CXCL10, chemokine ligand 10; DOXO, doxorubicin; ENO1, enolase-1; ENO2, enolase-2; ENO3, enolase-3; FOS, FOS proto-oncogene, AP-1 transcription factor subunit; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPI, glucose phosphate isomerase; Gy, gray; IFNs, interferons; IFI27, interferon alpha inducible protein 27; IFI44, interferon-induced protein 44; IFI44L, interferon-induced protein 44 like; IIFI6, interferon alpha inducible protein 6; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; IFIT3, interferon-induced protein with tetratricopeptide repeats 3; IFITM1, interferon-induced transmembrane protein 1; IRF7, interferon regulatory factor 7; ISG15, interferon-stimulated gene product 15; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; LDHA, lactate dehydrogenase A; LDHAL6B, lactate dehydrogenase A-like 6B; LDHB, lactate dehydrogenase B; M, myeloma; MCL-1, myeloid cell leukemia-1; MDH2, malate dehydrogenase 2; MM, metastatic melanoma; MX1, MX dynamin like GTPase 1; NPC, nasopharyngeal carcinoma; PGAM1, phosphoglycerate mutase 1; PGK1, phosphoglycerate kinase 1; OAS1, 2ʹ-5ʹ-Oligoadenylate synthetase 1; OASL, 2ʹ-5ʹ-Oligoadenylate synthetase like; PGM1, phosphoglucomutase 1; PKM2, pyruvate kinase muscle isozyme M2; γ-RAD, gamma radiation; SCC, squamous cell carcinoma; SDHA, succinate dehydrogenase complex, subunit A; TCT, tamoxifen; TPI1, triose phosphate isomerase 1; USP18, ubiquitin-specific peptidase 18.
Intrinsic Determinants in Tumor Radioresistance
Many in vivo and in vitro studies have demonstrated the critical roles of STAT1 in the induction of cell apoptosis.9–12 The “Friend” role of STAT1 in triggering apoptosis is modulated transcriptionally by anti-proliferation and pro-apoptotic signaling. Caspases, FAS, and TRAIL are the most well-known examples of genes controlled by STAT1 in these types of signaling.27 However, as mentioned above, high levels of constitutively active STAT1 are indispensable to support cell survival in several different types of cancer after RT. Table 1 presents a wide spectrum of multifunctional genes upregulated by STAT1 in different radioresistant tumor cells. Most of these radioresistance-associated genes are involved in anti-viral responses, DNA damage-induced apoptosis, and regulation of proliferation in the resistance to both X- and γ-radiation therapies. The activation of STAT1 by radiation may lead to tumor adaptation by directly affecting cellular signaling of the tumor cells.17 The cellular signaling triggered by IR occurs at two distinct sites: the nucleus and the cytoplasm. Intrinsic tumor radioresistance depends on the balance between DNA damage and DNA repair in the nucleus following irradiation.47 However, cytoplasmic signaling events, such as proliferation, anti-apoptosis, and pro-survival reaction, are equally important as DNA-related activities in the nucleus of radioresistant tumors. Therefore, the nuclear and cytoplasmic signaling pathways triggered by IR are described separately in this article in more detail.
Nuclear Events: The Balance Between DNA Damage Resistance and Apoptosis
IR leads to double-strand DNA breaks. Thus, protecting DNA from radiation-induced damage is one of the most important mechanisms tumor cells employ to develop RT resistance. Emerging evidence has shown that the constitutive expression of STAT1 and ISGs was associated with tumor DNA-damage resistance. In this regard, previous study reported that IFI27, IFI35, BST2, MX1, ISG15, and THBS1 were highly expressed in small cell lung cancer cells. These ISGs activated resistance to epigenetic modulators and other DNA damaging agents.48 Furthermore, there is a set of STAT1-induced genes, termed interferon-related DNA damage resistance signature (IRDS) genes, including IFI44, IFIT1, IFITM1, OAS1, ISG15, and MX1.12 These IRDS genes were derived from a primary breast cancer gene-expression profile that was more broadly associated with resistance to X-ray-induced DNA damage and doxorubicin.12 It should be noted that IRDS (+) breast cancer patients have a high rate of local-regional failure after adjuvant RT, which highlights the significance of IRDS in predicting recurrence after RT.12 The overexpression of the IRDS gene in glioblastoma multiforme (GBM) patients is related to poor survival outcome and RT response.49 Therefore, the balance of the STAT1-dependent genes involved in resistance to DNA damage is a critical determinant for the biological function of the IFN/STAT1 pathway in radioresistant cancer cells (the “Foe” role of STAT1). The purpose of these ISGs is mediated by the following factors: i) Different TME: the strong predictive value of IFI44, OAS1, ISG15, and MX1 in GBM patients was highly subtype-dependent and that the expression level of these ISGs was different in vitro and in vivo.49,50 ii) Manner of RT delivery: in breast cancer, prostate cancer, and gliosarcoma patients treated with multi-fraction radiation treatment (5 × 2 Gy) and a single-dose radiation treatment had different expression levels of both STAT1 and ISGs.50 iii) Modifications of STAT1: Numerous epigenetics and protein modifications can alter the transcriptional activity of STAT1 and potentially change the pattern of IFN/STAT1 pathway genes.17 Several studies have reported that the unphosphorylated STAT1 prolong the expression of ISGs, such as DDX60, G1P2, IFI27, IFI44, IFIT1, OAS1, OAS3, and PLSCR1. Most of their ISGs are upregulated in DNA damage-resistant cancer cells.51 In the contrary, acetylation of STAT1 counteracts IFN-induced STAT1 phosphorylation, nuclear translocation, and DNA binding.52 Moreover, acetylation of STAT1 also confers susceptibility to apoptosis by mediating p65-binding and suppressing anti-apoptotic NF-kB target genes in melanoma cells.53 Liu and colleagues demonstrate that Type 1 IFN signaling stimulates the translocation of USP12 from cytoplasm to the nucleus, that blocks the acetylation effect of CREB-binding protein (CBP) in nucleus.54 Therefore, in radioresistant cancer cells, aberrant IFNs signaling reduces the acetylation of STAT1 and inhibits the dephosphorylation of STAT1, leading to the high expression of STAT1.53,54 Consequently, balance between dynamic phosphorylation and acetylation of IFN-dependent STAT1 signaling regulates cancer response to RT. The interactions of STAT1 and ISGs’ roles in radiation-induced cell death are still unknown in various cancers. Hence, further research is needed for better understanding of the delicate regulation of STAT1-mediated acetylation and IFN signaling to overcome radioresistance in cancer.
Cytoplasmic Events: Pro-Survival Signaling
In contrast to nuclear signaling, radiation-induced cytoplasmic signaling events are much more likely to prompt a pro-survival response in radioresistant cancer cells.47 The efficacy of STAT1 to cause cell death and growth arrest depends on the transcriptional activation of pro-apoptotic genes and the inhibition of anti-apoptotic genes.27 In STAT1-overexpressing nu61 cancer cells, an impaired response to RT was observed.55 The expression of growth factor IL-6 and IL-8 in nu61 cells was higher relative to radiosensitive cells and knockdown of STAT1 led to the downregulation of these cytokines.55 Thus, a high level of STAT1 may be more likely to perform anti-apoptotic functions in radioresistant tumors (Figure 2). One hypothesis for this phenomenon is that under certain conditions, an RT-activated IFN/STAT1 pathway with pro-survival activity might antagonize apoptotic activity. For example, two IFN-induced genes, G1P3 with pro-survival activity and TRAIL with pro-apoptotic activity, antagonize each other in human myeloma cells.56 The other important pro-survival genes regulated by STAT1 signaling MCL-1,57 IFITM1,58 and USP18.59 Moreover, the dual function of STAT1 signaling depends on the cellular mechanisms, such as enzymes activation, glycolysis, oxidative phosphorylation, and mucin 1 oncoprotein, which is mainly expressed in human breast cancers, cooperatively worked with STAT1, indicating that STAT1-dependent genes in the context of STAT1 could predict a poor prognosis in breast cancers.60 In colon cancer cells, the cancer-upregulated gene 2 increased STAT1 activity to enhance cancer cell survival and migration.61 Likewise, in human head and neck squamous cell carcinoma cells, the active form of STAT1 was reported as a main control point in the X-ray suppression of the energy-related pathway.62 Briefly, STAT1, by upregulating many metabolism-associated genes, is involved in tumor glycolysis, oxidative phosphorylation, and the citrate cycle pathway (Table 1).62 As a result, the constitutive expression of STAT1 and STAT1-dependent genes may promote a switch from blocking the cytotoxic response to pro-tumor survival in radioresistant cancer cells.
MicroRNA: A Potential Radioresistance Regulator
MicroRNAs (miRNAs) are a class of small non-coding RNA approximately 22 nucleotides in length, known as critical post-transcriptional regulators, which regulate gene expression by binding to 3ʹ untranslated regions of the target mRNA.63 Recently, mechanistic studies from a range of tumor-bearing mouse models demonstrated some miRNAs involved in the STAT1-activated cellular process that were associated with the IR response by targeting the IFN/STAT1 signaling pathway. The miRNAs in Table 2 act as tumor-promoter and tumor-suppressor miRNAs by modulating the oncogenic function of STAT1 in different tumor contexts. Consequently, it shows the complexity of the IFN/STAT1-miRNAs regulatory network in cancer cells to stimulate the STAT1 positive feedback loop and STAT1-dependent gene expression. Remarkably, the balance between IFN/STAT1 and tumor-promoting miRNAs (oncomiRNAs) promotes the epithelial-to-mesenchymal transition process, increasing radioresistance and perhaps, distant metastasis of tumors.64 Besides that, the feedback relationship between STAT1-targeted miRNAs, such as miR-99a,65 miR-20366 and STAT1-dependent genes, is likely to regulate STAT1 signaling at the TME level (Figure 1). Thus, blocking oncomiRNAs or activating tumor suppressor miRNAs may be effective ways to improve tumor radiosensitivity (Table 2).
Table 2.
MicroRNA Regulate the IFNs/STAT1 Signaling Pathway in Cancer
| Name | Function | Role on INFs/JAK/STAT1 | Cancer | References |
|---|---|---|---|---|
| Tumor Suppressor miRNAs | ||||
| miR-15A miR-16-1 |
Inhibit immunosuppressive B cells accumulation and repress of tumor growth | Inhibit activation of STAT1 | CRC | [85] |
| miR-99a | Induce apoptosis and inhibit proliferation | Downregulate the phosphorylation levels of STAT1 | Rb | [65] |
| miR-203 | Inhibit cell proliferation and migration and increase sensitivity to apoptosis | Inhibit activation of STAT1 | GBM | [66] |
| miR-375 | Inhibit PD-L1 expression | Inhibit JAK2/STAT1 signaling | HNSCC | [86] |
| miR-944 | Inhibit tumor cell growth and proliferation | Downregulate expression of STAT1 | LAC | [87] |
| oncomiRNAs* | ||||
| miR-9 | Promotes endothelial cell migration and angiogenesis | Activate oncogenic function of STAT1 | NSCLC, M, PC, GBM, CRC | [88] |
| miR-19b miR-30b |
Negative regulation of SOCS1 and SOCS3 | Regulate constant overexpression of STAT1 | MPM | [89] |
| miR-30c miR-2909 |
Contributes to cell growth | Activate oncogenic function of STAT1 | PCa | [90] |
| miR-101 miR-128 miR-363 |
Interaction with IFIT5 and reduce cancer metastasis | Regulate IFN-γ-JAK/STAT1 induced EMT | PCa | [64] |
| miR-221 miR-222 |
Regulate IRDS expression contribute to radioresistance | Regulate STAT1 tyrosine phosphorylation | Glioma | [17, 91] |
Note: *Micro-RNAs are involved in tumorigenesis.
Abbreviations: CRC, colorectal cancer; EMT, epithelial-to-mesenchymal transition; GBM, glioblastoma; HNSCC, head and neck squamous cell carcinoma; JAK, Janus kinase; IFIT5, interferon-induced tetratricopeptide repeat 5; IFN, interferon; IFN-γ, interferon-γ; IRDS, interferon-related DNA damage resistance signature; LAC, lung adenocarcinoma; LLC1, Lewis lung carcinoma; M, myeloma; miR; micro-RNA; MPM, malignant pleural mesothelioma; NSCLC, non-small cell lung cancer; PC, pancreatic cancer; PCa, prostate cancer; PD-L1, programmed-death ligand 1; RB, retinoblastoma; RCC, renal cell carcinoma; STAT1, signal transducer and activator of transcription 1; SOCS1, suppressor of cytokine signaling 1; SOCS3, suppressor of cytokine signaling 3; SOCS5, suppressor of cytokine signaling 5.
IFN/STAT1 Signaling Pathway in Drug Resistance
Similar to that observed in RT, chronic exposure to both Type I and Type II IFNs may also lead to the STAT1-mediated mechanism of drug resistance. A correlation between STAT1 expression and cisplatin resistance in human ovarian cancer cells has been reported in many in vivo and in vitro studies. As shown in Table 1, cisplatin-, tamoxifen- or doxorubicin-resistant cancer cells demonstrated the increased expression of different ISGs, including DDX60, OAS1, G1P3, and IFI27.67 The STAT1-dependent association with histone deacetylase (HDAC)-4 axis enhances the acquired platinum resistance in ovarian cancer cells, and also has been linked to the failure of etoposide in lung cancer cells.68,69 Also, in a prostate cancer cell line, DU145-DR, STAT1 and its subsequent regulation of clustering genes, was essential for docetaxel resistance.70 Moreover, prolonged STAT1 signaling can mediate tumor cell resistance to both RT and chemotherapy. As mentioned above, cancers with high expressions of IRDS genes are cross-resistant to doxorubicin and RT (Figure 1).12 Intriguingly, combination therapy of Type II IFN with doxorubicin in MDA-MB 435 breast cancer cells activated STAT1 and enhanced doxorubicin-induced apoptosis.71 Therefore, tumor cells, with the participation of IR, may develop another resistant mechanism mediated by the upregulation of STAT1 and/or STAT1-induced genes, resulting in an impaired anti-tumor IFN/STAT1 signaling response. The role of STAT1 signaling response in combined RT and chemotherapy is complex and driven by the expression of many subsets of ISGs in different cancer. Further explorations are required to unveil how STAT1-mediates cross-resistance to RT and chemotherapy and to confirm that targeting STAT1 may be an effective therapeutic technique to improve the effectiveness of anti-cancer treatment.
Future Direction
Constitutively activated STATs, particularly STAT1, STAT3, and STAT5 have been found in a variety of human tumors. Many preclinical trials aim to improve the effectiveness of anti-tumor therapy by stimulating the anti-neoplastic function of STAT1 and inhibiting the oncogenic function of STAT3 and STAT5.7,72 Despite extensive studies on STATs activation and its role in RT, still STAT1 and STAT3 represent an attractive target in the modern era of radio-immunotherapy.8
Although STAT3 is of great interest and the deeply investigated STAT protein in cancer research, emerging findings suggest STAT1 as a critical mediator of oncogenic signaling and participate in the formation of radioresistant phenotype in some cancers, such as breast cancer,12 head and neck cancer,9 myeloma,10 and renal cell carcinoma.11 In these tumor cells with high expression of STAT1, targeting IFN/STAT1 pathway may lead to the discovery of new treatment strategies for targeted RT. Undoubtedly, the potential usefulness of STAT-targeted therapeutics in RT needs more investigation to unravel the relevant physiological pathway of the STAT protein family. In addition, the possibility of the combination of STATs inhibitors with other targeted therapy will be useful in the synthetic lethality approach of RT; which depends on the availability of a clinically relevant delivery system. While the mechanisms of cancer radioresistance have not been fully elucidated, the results of previous studies suggest that the outcome of IFN/STAT1 pathway effects in tumors after radiation depends on the intensity of the signal, microenvironment cues, and the tumor-specific context.5 Therefore, strategies to overcome resistance in the context of the IFN/STAT1 pathway need to take into full account how to balance the pro-survival and cytotoxic aspects of STAT1 signaling in RT (Figure 2). IFN/STAT1 approaches to reduce tumor radioresistance have been derived from new findings and have not yet been used in clinical settings.18 Treatments targeting STAT1 may be verified as radiosensitizing strategies in various pre-clinical settings. Future comprehensive molecular and cellular investigations need to focus on the roles of the IFN/STAT1 pathway in radioresistant cancer cells. IFN/STAT1 pathway blockers can be divided into the following: i) targeting the JAK/STAT1 signaling. Several JAK inhibitors have already received FDA approval for rheumatoid arthritis, including the upadacitinib,73 baricitinib,74 and tofacitinib.75 Besides, fludarabine has been described as a radiosensitizer, by blocking STAT1 improves the efficacy of RT in many cancers.76,77 Moreover, some preclinical studies have found that targeting IFN/STAT1 signaling effectively reduces the expression of PD-L1 in cancer cells. For example, SAR302503, a selected JAK2 inhibitor, has been reported to suppress STAT1 activation in radioresistant non-small cell lung cancer (NSCLC) cell lines, as well as abrogate tumor cell-intrinsic expression of PD-L1 in NSCLC cells.78 Also, 15-deoxy-δ12, 14-prostaglandin J2, a peroxisome proliferator-activated receptor-gamma activator, can strongly downregulate Type II IFN-elicited PD-L1 expression through targeting the JAK/STAT1 signaling pathway in melanoma cells.79 In this field, efficacy in appropriate delivery of IFN/STAT1 signaling inhibitor, biodistribution, selectivity and preliminary biosafety are the major challenges for pharmacologists; ii) targeting STAT1 modulation. As discussed above, acetylation of STAT1 inhibits IFN-dependent STAT1 phosphorylation and STAT1-dependent gene expression pattern.68 The upregulation of the acetylation of Type II IFN/STAT1 pathways in cancer cells may improve the sensitivity to RT. Acetylation of STAT1 correlates with the dissociation of HDACs and interaction with CBP.52 In cisplatin-resistant ovarian cancer cells, silencing of HDAC-4 increases the STAT1 acetylation levels, and restores cisplatin sensitivity.68 Thus, targeting the phosphorylation-acetylation switch may be a promising strategy to regulate STAT1 signaling. (iii) targeting the STAT1-activated genes. As Table 1 shows, some STAT1-induced genes, especially ISGs, can not only mediate resistance to radiation but also mediate resistance to some anti-neoplastic drugs, such as doxorubicin and cisplatin. Certainly, approaches to target these genes in radioresistant cancer cells may have significant benefits in cancer patients undergoing RT. However, considering the original function of these drugs, such as anti-viral activity, warrant further investigation in the clinic. (iv) targeting the miRNAs. Suppressing or inhibiting oncomiRNAs is a potential method to impair the oncogenic function of the IFN/STAT1 pathway. Additionally, activating of tumor-suppressor miRNAs, may increase the activation state of STAT1 in radioresistant tumors and improve the tumor radiosensitivity. (v) targeting the STAT3 protein. It is well established that STAT1 and STAT3 can counteract each other’s functions by competing for the same DNA binding sequence.8 However, positive association between levels of STAT1 and STAT3 has been found in breast cancer cells.80 Through binding to the STAT1 promoter, STAT3 can significantly enhance STAT1 gene expression, as well as activates the expression of iNOS and COX-2.80 Because the homology between STAT1 and STAT3, off-target STAT1 blockade by a STAT3 inhibitor may result in some benefit effects in radioresistant cancers.8,81 For example, Honokiol, a novel blocker of STAT3 activation, has a great potential for the treatment of hepatocellular carcinoma, that also suppresses constitutive activation of JAK1 and JAK2.16 Of note, the ratio of STAT1 and STAT3 expression constitutes important mechanisms in controlling their response to cytokines and growth factors.72 Thus, targeting STAT3 therapy should take into full account about the balance state of STAT1 and STAT3, as well as reduce its toxic and adverse effects to achieve more stable efficacy.
Conclusions
In conclusion, this review highlights the mechanistic functions of the IFN/STAT1 pathway to confer radioresistance and radiosensitization in radio-immunotherapy, which require further comprehensive studies. The possibility of a radiation-induced IFN/STAT1 mechanism will open new opportunities for the development of novel therapeutic strategies, as well as define a new approach to enhance and radio-immunotherapy efficacy in the clinical treatment of various cancers. In the future, more investigations are needed into the interplay between the overexpression of STAT1 signaling and STAT1-dependent genes in different tumors. Meanwhile, laboratory investigations need to continue to elucidate the paradoxical effects of the IFN/STAT1 pathway response to RT.
Acknowledgments
We gratefully acknowledge all authors and coworkers for their contributions. This work was supported by grants from the Union Project of Luzhou City and the Southwest Medical University (Nos. 14JC0144, 2013LZLY-J40).
Abbreviations
CBP, CREB-binding protein; CSCs, cancer stem cells; GBM, glioblastoma multiforme; HDAC, histone deacetylase; IDO, indoleamine-2,3-dioxygenase; IFNs, interferons; IFITs, interferon-induced protein with tetratricopeptide repeats; iNOS, inducible nitric oxide synthase; IRDS, interferon-related DNA damage resistance signature; IR, ionizing radiation; ISGs, interferon-stimulated genes; ISGF3, interferon-stimulated gene factor 3; JAK, Janus kinase; MDM2, murine double minute 2; MDSCs, myeloid-derived suppressor cells; miRNAs, microRNAs; MX1, interferon-induced GTP-binding protein; NSCLC, non-small cell lung cancer, PD-L1, programmed death-ligand 1; RT, radiation therapy; SERPINB9, serpin family B member 9; STATs, signal transducer and activator of transcriptions; TAMs, tumor-associated macrophages; TME, tumor microenvironment; Tregs, regulatory T-cells.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31(4):363–372. doi: 10.1007/s13277-010-0042-8 [DOI] [PubMed] [Google Scholar]
- 2.Herrera FG, Bourhis J, Coukos G. Radiotherapy combination opportunities leveraging immunity for the next oncology practice. CA Cancer J Clin. 2017;67(1):65–85. doi: 10.3322/caac.21358 [DOI] [PubMed] [Google Scholar]
- 3.Darragh LB, Oweida AJ, Karam SD. Overcoming resistance to combination radiation-immunotherapy: a focus on contributing pathways within the tumor microenvironment. Front Immunol. 2018;9:3154. doi: 10.3389/fimmu.2018.03154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15(7):409–425. doi: 10.1038/nrc3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018;9(847):1–19. doi: 10.3389/fimmu.2018.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugade AA, Sorensen EW, Gerber SA, Moran JP, Frelinger JG, Lord EM. Radiation-induced IFN-gamma production within the tumor microenvironment influences antitumor immunity. J Immunol. 2008;180(5):3132–3139. doi: 10.4049/jimmunol.180.5.3132 [DOI] [PubMed] [Google Scholar]
- 7.Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double-edged sword of immune regulation and cancer progression. Cancers. 2019;11(12):2002. doi: 10.3390/cancers11122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhoeven Y, Tilborghs S, Jacobs J, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol. 2020;60:41–56. doi: 10.1016/j.semcancer.2019.10.002 [DOI] [PubMed] [Google Scholar]
- 9.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci U S A. 2004;101(6):1714–1719. doi: 10.1073/pnas.0308102100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryknas M, Dhar S, Oberg F, et al. STAT1 signaling is associated with acquired crossresistance to doxorubicin and radiation in myeloma cell lines. Int J Cancer. 2007;120(1):189–195. doi: 10.1002/ijc.22291 [DOI] [PubMed] [Google Scholar]
- 11.Hui Z, Tretiakova M, Zhang Z, et al. Radiosensitization by inhibiting STAT1 in renal cell carcinoma. Int J Radiat Oncol Biol Phys. 2009;73(1):288–295. doi: 10.1016/j.ijrobp.2008.08.043 [DOI] [PubMed] [Google Scholar]
- 12.Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S, Guo Y, Huang ST, Zhu XD. Inhibition of STAT1 sensitizes radioresistant nasopharyngeal carcinoma cell line CNE-2R to radiotherapy. Oncotarget. 2018;9(9):8303–8310. doi: 10.18632/oncotarget.19690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Wang Y, Teng M, Zhang D, Li L, Liu Y. Signal transducers and activators of transcription-1 (STAT1) regulates microRNA transcription in interferon gamma-stimulated HeLa cells. PLoS One. 2010;5(7):1–8. doi: 10.1371/journal.pone.0011794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang SH, Li L, Xie YQ, et al. IFN-gamma-STAT1-iNOS induces myeloid progenitors to acquire immunosuppressive activity. Front Immunol. 2017;8(1192):1–12. doi: 10.3389/fimmu.2017.01192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajendran P, Li F, Shanmugam MK, et al. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J Cell Physiol. 2012;227(5):2184–2195. doi: 10.1002/jcp.22954 [DOI] [PubMed] [Google Scholar]
- 17.Khodarev NN, Roizman B, Weichselbaum RR. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin Cancer Res. 2012;18(11):3015–3021. doi: 10.1158/1078-0432.ccr-11-3225 [DOI] [PubMed] [Google Scholar]
- 18.Budhwani M, Mazzieri R, Dolcetti R. Plasticity of type I interferon-mediated responses in cancer therapy: from anti-tumor immunity to resistance. Front Oncol. 2018;8(322):1–16. doi: 10.3389/fonc.2018.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res. 2011;17(19):6118–6124. doi: 10.1158/1078-0432.ccr-11-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seliger B, Ruiz-Cabello F, Garrido F. IFN inducibility of major histocompatibility antigens in tumors. Adv Cancer Res. 2008;101(1):1–28. doi: 10.1016/s0065-230x(08)00407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Cao Y, Markelc B, Kaeppler J, Vermeer JA, Muschel RJ. Type I IFN protects cancer cells from CD8+ T cell-mediated cytotoxicity after radiation. J Clin Invest. 2019;129(10):4224–4238. doi: 10.1172/jci127458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17(8):457–474. doi: 10.1038/nrc.2017.51 [DOI] [PubMed] [Google Scholar]
- 23.Zheng P, Sarma S, Guo Y, Liu Y. Two mechanisms for tumor evasion of preexisting cytotoxic T-cell responses: lessons from recurrent tumors. Cancer Res. 1999;59(14):3461–3467. [PubMed] [Google Scholar]
- 24.Juneja VR, McGuire KA, Manguso RT, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214(4):895–904. doi: 10.1084/jem.20160801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Townsend PA, Scarabelli TM, Davidson SM, Knight RA, Latchman DS, Stephanou A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J Biol Chem. 2004;279(7):5811–5820. doi: 10.1074/jbc.M302637200 [DOI] [PubMed] [Google Scholar]
- 26.Qin JJ, Li X, Hunt C, Wang W, Wang H, Zhang R. Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine. Genes Dis. 2018;5(3):204–219. doi: 10.1016/j.gendis.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24(130):19–29. [PubMed] [Google Scholar]
- 28.Moon JW, Kong SK, Kim BS, et al. IFNgamma induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep. 2017;7(1):17810. doi: 10.1038/s41598-017-18132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morimoto Y, Kishida T, Kotani SI, Takayama K, Mazda O. Interferon-beta signal may up-regulate PD-L1 expression through IRF9-dependent and independent pathways in lung cancer cells. Biochem Biophys Res Commun. 2018;507(1–4):330–336. doi: 10.1016/j.bbrc.2018.11.035 [DOI] [PubMed] [Google Scholar]
- 30.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4(5):336–347. doi: 10.1038/nri1349 [DOI] [PubMed] [Google Scholar]
- 31.Bellucci R, Martin A, Bommarito D, et al. Interferon-gamma-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology. 2015;4(6):1–10. doi: 10.1080/2162402x.2015.1008824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benci JL, Xu B, Qiu Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554 e1512. doi: 10.1016/j.cell.2016.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J Immunol. 2000;164(7):3596–3599. doi: 10.4049/jimmunol.164.7.3596 [DOI] [PubMed] [Google Scholar]
- 34.Mellor AL, Keskin DB, Johnson T, Chandler P, Munn DH. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J Immunol. 2002;168(8):3771–3776. doi: 10.4049/jimmunol.168.8.3771 [DOI] [PubMed] [Google Scholar]
- 35.Monjazeb AM, Kent MS, Grossenbacher SK, et al. Blocking indolamine-2,3-dioxygenase rebound immune suppression boosts antitumor effects of radio-immunotherapy in murine models and spontaneous canine malignancies. Clin Cancer Res. 2016;22(17):4328–4340. doi: 10.1158/1078-0432.ccr-15-3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achyut BR, Arbab AS. Myeloid cell signatures in tumor microenvironment predicts therapeutic response in cancer. Onco Targets Ther. 2016;9:1047–1055. doi: 10.2147/ott.s102907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albini A, Bruno A, Noonan DM, Mortara L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment: implications for immunotherapy. Front Immunol. 2018;9:527. doi: 10.3389/fimmu.2018.00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hix LM, Karavitis J, Khan MW, Shi YH, Khazaie K, Zhang M. Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. J Biol Chem. 2013;288(17):11676–11688. doi: 10.1074/jbc.M112.441402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol. 2005;174(8):4880–4891. doi: 10.4049/jimmunol.174.8.4880 [DOI] [PubMed] [Google Scholar]
- 40.Wei B, Baker S, Wieckiewicz J, Wood KJ. IFN-gamma triggered STAT1-PKB/AKT signalling pathway influences the function of alloantigen reactive regulatory T cells. Am J Transplant. 2010;10(1):69–80. doi: 10.1111/j.1600-6143.2009.02858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart CA, Metheny H, Iida N, et al. Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J Clin Invest. 2013;123(11):4859–4874. doi: 10.1172/jci65180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brody JR, Costantino CL, Berger AC, et al. Expression of indoleamine 2,3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle. 2009;8(12):1930–1934. doi: 10.4161/cc.8.12.8745 [DOI] [PubMed] [Google Scholar]
- 43.Schulz A, Meyer F, Dubrovska A, Borgmann K. Cancer stem cells and radioresistance: DNA repair and beyond. Cancers. 2019;11(862):1–20. doi: 10.3390/cancers11060862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qadir AS, Ceppi P, Brockway S, et al. CD95/Fas increases stemness in cancer cells by inducing a STAT1-dependent type I interferon response. Cell Rep. 2017;18(10):2373–2386. doi: 10.1016/j.celrep.2017.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhan JF, Chen LH, Yuan YW, et al. STAT1 promotes radioresistance of CD44(+)/CD24(-/low) cells in breast cancer. Exp Biol Med (Maywood). 2011;236(4):418–422. doi: 10.1258/ebm.2011.010287 [DOI] [PubMed] [Google Scholar]
- 46.Wang C, Zheng W, Yao D, et al. Upregulation of DNA metabolism-related genes contributes to radioresistance of glioblastoma. Hum Gene Ther Clin Dev. 2019;00(251):1–14. doi: 10.1089/humc.2018.251 [DOI] [PubMed] [Google Scholar]
- 47.Deorukhkar A, Krishnan S. Targeting inflammatory pathways for tumor radiosensitization. Biochem Pharmacol. 2010;80(12):1904–1914. doi: 10.1016/j.bcp.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luszczek W, Cheriyath V, Mekhail TM, Borden EC. Combinations of DNA methyltransferase and histone deacetylase inhibitors induce DNA damage in small cell lung cancer cells: correlation of resistance with IFN-stimulated gene expression. Mol Cancer Ther. 2010;9(8):2309–2321. doi: 10.1158/1535-7163.mct-10-0309 [DOI] [PubMed] [Google Scholar]
- 49.Duarte CW, Willey CD, Zhi D, et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One. 2012;7(1):1–8. doi: 10.1371/journal.pone.0029653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai MH, Cook JA, Chandramouli GV, et al. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67(8):3845–3852. doi: 10.1158/0008-5472.can-06-4250 [DOI] [PubMed] [Google Scholar]
- 51.Cheon H, Holvey-Bates EG, Schoggins JW, et al. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32(20):2751–2763. doi: 10.1038/emboj.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krämer OH, Knauer SK, Greiner G, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23(2):223–235. doi: 10.1101/gad.479209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krämer OH, Baus D, Knauer SK, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20(4):473–485. doi: 10.1101/gad.364306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Jin L, Chen X, et al. USP12 translocation maintains interferon antiviral efficacy by inhibiting CBP acetyltransferase activity. PLoS Pathog. 2020;16(1):e1008215. doi: 10.1371/journal.ppat.1008215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efimova EV, Liang H, Pitroda SP, et al. Radioresistance of Stat1 over-expressing tumour cells is associated with suppressed apoptotic response to cytotoxic agents and increased IL6-IL8 signalling. Int J Radiat Biol. 2009;85(5):421–431. doi: 10.1080/09553000902838566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheriyath V, Glaser KB, Waring JF, Baz R, Hussein MA, Borden EC. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest. 2007;117(10):3107–3117. doi: 10.1172/jci31122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timofeeva OA, Plisov S, Evseev AA, et al. Serine-phosphorylated STAT1 is a prosurvival factor in Wilms’ tumor pathogenesis. Oncogene. 2006;25(58):7555–7564. doi: 10.1038/sj.onc.1209742 [DOI] [PubMed] [Google Scholar]
- 58.Kita K, Sugaya S, Zhai L, et al. Involvement of LEU13 in interferon-induced refractoriness of human RSa cells to cell killing by X rays. Radiat Res. 2003;160(3):302–308. doi: 10.1667/RR3039 [DOI] [PubMed] [Google Scholar]
- 59.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70(2):655–665. doi: 10.1158/0008-5472.can-09-1942 [DOI] [PubMed] [Google Scholar]
- 60.Khodarev N, Ahmad R, Rajabi H, et al. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2010;29(6):920–929. doi: 10.1038/onc.2009.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malilas W, Koh SS, Kim S, et al. Cancer upregulated gene 2, a novel oncogene, enhances migration and drug resistance of colon cancer cells via STAT1 activation. Int J Oncol. 2013;43(4):1111–1116. doi: 10.3892/ijo.2013.2049 [DOI] [PubMed] [Google Scholar]
- 62.Pitroda SP, Wakim BT, Sood RF, et al. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 2009;7(68):1–10. doi: 10.1186/1741-7015-7-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imani S, Wu RC, Fu J. MicroRNA-34 family in breast cancer: from research to therapeutic potential. J Cancer. 2018;9(20):3765–3775. doi: 10.7150/jca.25576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo UG, Pong RC, Yang D, et al. IFNgamma-induced IFIT5 promotes epithelial-to-mesenchymal transition in prostate cancer via miRNA processing. Cancer Res. 2019;79(6):1098–1112. doi: 10.1158/0008-5472.can-18-2207 [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Sun W, Han N, Zou Y, Yin D. Curcumin inhibits proliferation, migration, invasion and promotes apoptosis of retinoblastoma cell lines through modulation of miR-99a and JAK/STAT pathway. BMC Cancer. 2018;18(1):1–9. doi: 10.1186/s12885-018-5130-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Yang CH, Wang Y, Sims M, et al. MiRNA203 suppresses the expression of protumorigenic STAT1 in glioblastoma to inhibit tumorigenesis. Oncotarget. 2016;7(51):84017–84029. doi: 10.18632/oncotarget.12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Post AEM, Smid M, Nagelkerke A, et al. Interferon-stimulated genes are involved in cross-resistance to radiotherapy in tamoxifen-resistant breast cancer. Clin Cancer Res. 2018;24(14):3397–3408. doi: 10.1158/1078-0432.ccr-17-2551 [DOI] [PubMed] [Google Scholar]
- 68.Stronach EA, Alfraidi A, Rama N, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 2011;71(13):4412–4422. doi: 10.1158/0008-5472.can-10-4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaewpiboon C, Srisuttee R, Malilas W, et al. Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol Med Rep. 2015;11(3):2315–2321. doi: 10.3892/mmr.2014.2949 [DOI] [PubMed] [Google Scholar]
- 70.Patterson SG, Wei S, Chen X, et al. Novel role of Stat1 in the development of docetaxel resistance in prostate tumor cells. Oncogene. 2006;25(45):6113–6122. doi: 10.1038/sj.onc.1209632 [DOI] [PubMed] [Google Scholar]
- 71.Thomas M, Finnegan CE, Rogers KM, et al. STAT1: a modulator of chemotherapy-induced apoptosis. Cancer Res. 2004;64(22):8357–8364. doi: 10.1158/0008-5472.can-04-1864 [DOI] [PubMed] [Google Scholar]
- 72.Wong ALA, Hirpara JL, Pervaiz S, Eu JQ, Sethi G, Goh BC. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin Investig Drugs. 2017;26(8):883–887. doi: 10.1080/13543784.2017.1351941 [DOI] [PubMed] [Google Scholar]
- 73.Smolen JS, Pangan AL, Emery P, et al. Upadacitinib as monotherapy in patients with active rheumatoid arthritis and inadequate response to methotrexate (SELECT-MONOTHERAPY): a randomised, placebo-controlled, double-blind Phase 3 study. Lancet (London, England). 2019;393(10188):2303–2311. doi: 10.1016/s0140-6736(19)30419-2 [DOI] [PubMed] [Google Scholar]
- 74.Mogul A, Corsi K, McAuliffe L. Baricitinib: the second FDA-approved JAK inhibitor for the treatment of rheumatoid arthritis. Ann Pharmacother. 2019;53(9):947–953. doi: 10.1177/1060028019839650 [DOI] [PubMed] [Google Scholar]
- 75.Dhillon S. Tofacitinib: a review in rheumatoid arthritis. Drugs. 2017;77(18):1987–2001. doi: 10.1007/s40265-017-0835-9 [DOI] [PubMed] [Google Scholar]
- 76.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5(4):444–447. doi: 10.1038/7445 [DOI] [PubMed] [Google Scholar]
- 77.Gregoire V, Hunter N, Milas L, Brock WA, Plunkett W, Hittelman WN. Potentiation of radiation-induced regrowth delay in murine tumors by fludarabine. Cancer Res. 1994;54(2):468–474. [PubMed] [Google Scholar]
- 78.Pitroda SP, Stack ME, Liu GF, et al. JAK2 inhibitor SAR302503 abrogates PD-L1 expression and targets therapy-resistant non-small cell lung cancers. Mol Cancer Ther. 2018;17(4):732–739. doi: 10.1158/1535-7163.mct-17-0667 [DOI] [PubMed] [Google Scholar]
- 79.Seo SK, Seo DI, Park WS, et al. Attenuation of IFN-gamma-induced B7-H1 expression by 15-deoxy-delta(12,14)-prostaglandin J2 via downregulation of the Jak/STAT/IRF-1 signaling pathway. Life Sci. 2014;112(1–2):82–89. doi: 10.1016/j.lfs.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 80.Han W, Carpenter RL, Cao X, Lo HW. STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation with STAT3. Mol Carcinog. 2013;52(12):959–969. doi: 10.1002/mc.21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Munoz J, Dhillon N, Janku F, Watowich SS, Hong DS. STAT3 inhibitors: finding a home in lymphoma and leukemia. Oncologist. 2014;19(5):536–544. doi: 10.1634/theoncologist.2013-0407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khodarev NN, Minn AJ, Efimova EV, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67(19):9214–9220. doi: 10.1158/0008-5472.can-07-1019 [DOI] [PubMed] [Google Scholar]
- 83.Khodarev NN, Roach P, Pitroda SP, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4(6):1–14. doi: 10.1371/journal.pone.0005821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo Y, Zhu XD, Qu S, et al. Identification of genes involved in radioresistance of nasopharyngeal carcinoma by integrating gene ontology and protein-protein interaction networks. Int J Oncol. 2012;40(1):85–92. doi: 10.3892/ijo.2011.1172 [DOI] [PubMed] [Google Scholar]
- 85.Liu R, Lu Z, Gu J, et al. MicroRNAs 15A and 16-1 activate signaling pathways that mediate chemotaxis of immune regulatory B cells to colorectal tumors. Gastroenterology. 2018;154(3):637–651e637. doi: 10.1053/j.gastro.2017.09.045 [DOI] [PubMed] [Google Scholar]
- 86.Wu Q, Zhao Y, Sun Y, Yan X, Wang P. miR-375 inhibits IFN-gamma-induced programmed death 1 ligand 1 surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol Rep. 2018;39(3):1461–1468. doi: 10.3892/or.2018.6177 [DOI] [PubMed] [Google Scholar]
- 87.An JC, Shi HB, Hao WB, Zhu K, Ma B. miR-944 inhibits lung adenocarcinoma tumorigenesis by targeting STAT1 interaction. Oncol Lett. 2019;17(4):3790–3798. doi: 10.3892/ol.2019.10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhuang G, Wu X, Jiang Z, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–3523. doi: 10.1038/emboj.2012.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arzt L, Halbwedl I, Gogg-Kamerer M, Popper HH. Signal transducer and activator of transcription 1 (STAT1) knock-down induces apoptosis in malignant pleural mesothelioma. Pathol Oncol Res. 2017;23(3):595–605. doi: 10.1007/s12253-016-0157-3 [DOI] [PubMed] [Google Scholar]
- 90.Ayub SG, Kaul D. miR-2909 regulates ISGylation system via STAT1 signalling through negative regulation of SOCS3 in prostate cancer. Andrology. 2017;5(4):790–797. doi: 10.1111/andr.12374 [DOI] [PubMed] [Google Scholar]
- 91.Zhang C, Han L, Zhang A, et al. Global changes of mRNA expression reveals an increased activity of the interferon-induced signal transducer and activator of transcription (STAT) pathway by repression of miR-221/222 in glioblastoma U251 cells. Int J Oncol. 2010;36(6):1503–1512. [DOI] [PubMed] [Google Scholar]