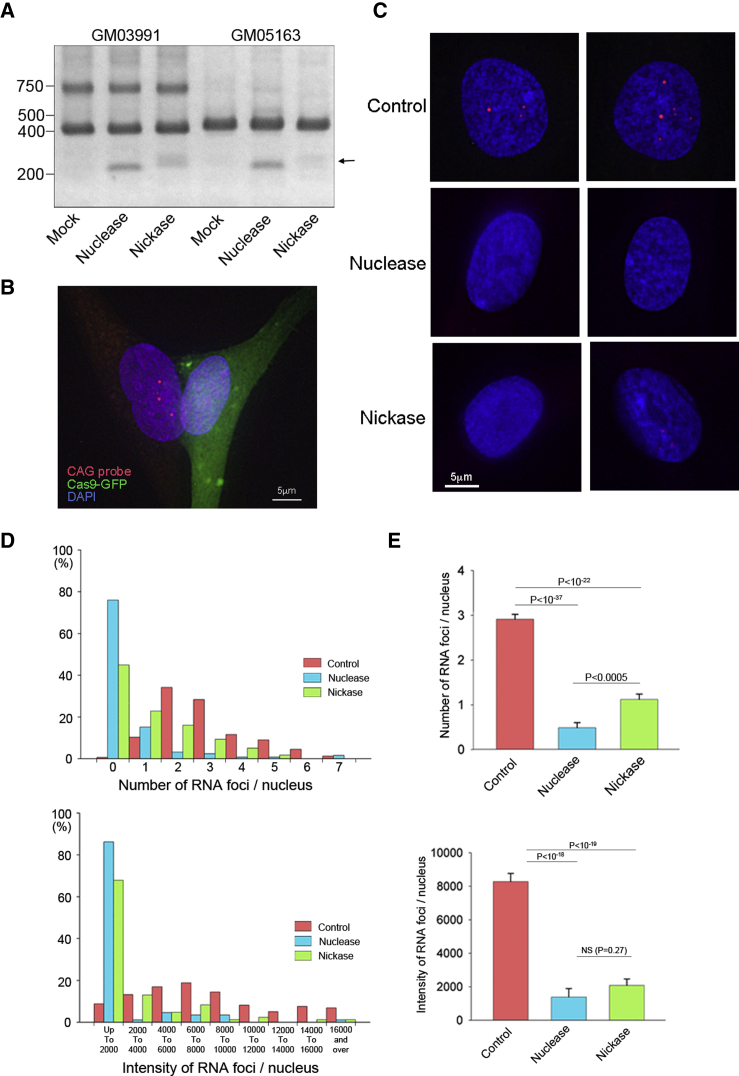
Figure 3.
Suppression of RNA Foci Formation by the Repeat Excision
(A) Fibroblast GM03991, harboring 50–80 CTG repeats, was found to consist of a wild-type allele at 400 bp and a mutant allele at 700 bp by PCR. However, fibroblast GM05163, with 400 CTG repeats, exhibited only the wild-type allele, presumably because the PCR failed to amplify the highly repetitive sequence of the mutant allele. Genome editing was performed to these cells using conventional Cas9 nuclease and two sgRNAs (5′ guide 3 and 3′ guide 2) or the double nicking strategy with Cas9 nickase and four sgRNAs (Nick 1 and Nick 3 pairs). Using both of the procedures, lower molecular weight bands were observed above 200 bp (arrow), indicating the successful excision of the CTG repeat. (B) Fibroblast GM05163 was co-transfected with GFP-tagged Cas9 nuclease or Cas9 nickase together with sgRNAs. The RNA-FISH image shows intense RNA foci in the GFP-negative fibroblast (left) but not in the GFP-positive Cas9 nuclease-expressing cell (right). (C) In the nuclei of the control fibroblasts (no plasmids), several RNA foci were consistently observed. However, the RNA foci were mostly undetectable in fibroblasts expressing Cas9 nuclease or nickase. (D) Quantitative analyses of the number and the total intensity of intranuclear RNA foci were performed. Histograms of both the number (upper panel) and the intensity (lower panel) with a left-side skew were obtained for Cas9 nuclease and nickase. (E) The average number of RNA foci was significantly reduced by both Cas9 nuclease and nickase. When compared between Cas9 nuclease and nickase, the nuclease-treated cells exhibited significantly fewer foci (upper panel). The average intensity of RNA foci was significantly decreased by both Cas9 nuclease and nickase. There was no significant difference between the nuclease and nickase by this parameter (lower panel). The results are expressed as the mean ± SEM.