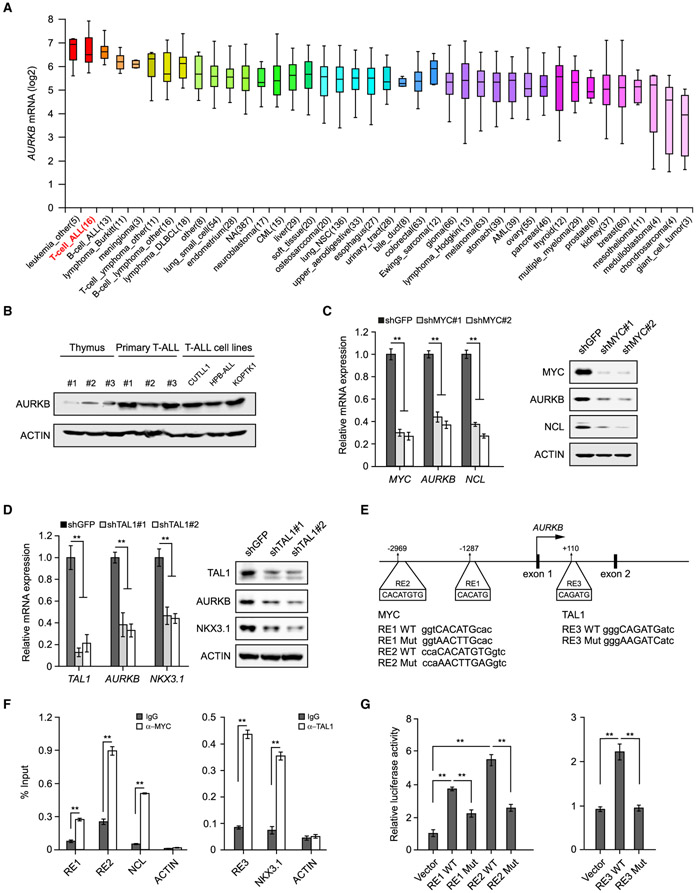
Figure 4. MYC and TAL1 Activate AURKB Transcription.
(A) AURKB mRNA expression was analyzed among 1,457 human cancer cell lines in CCLE database (https://portals.broadinstitute.org/ccle). The distributions of AURKB mRNA expression are presented as log2 median-centered intensity and shown in box-and-whisker plots, which depict the first and third quartiles, with the median shown as a solid line inside the box and whiskers extending to 1.5 interquartile range from first and third quartiles.
(B) AURKB proteins were analyzed by immunoblot in normal thymuses from healthy donors, primary T-ALL samples and T-ALL cell lines, respectively.
(C) AURKB mRNA and protein were analyzed in MYC-depleted CUTLL1 cells by qRT-PCR and immunoblot. MYC target gene NCL was used as a positive control.
(D) AURKB mRNA and protein were analyzed in TAL1-depleted Jurkat cells by qRT-PCR and immunoblot. TAL1 target gene NKX3.1 was used as a positive control.
(E) Schematic presentation of MYC and TAL1 binding sites on the AURKB locus. MYC response elements, RE1 and RE2; TAL1 responsive element, RE3. Consensus sequence mutations are shown as RE Mut.
(F) ChIP analysis of MYC binding to the AURKB locus in CUTLL1 cells (left) orTAL1 binding in Jurkat cells (right). Binding signals to NCL and NKX3.1 are shown as positive controls, ACTIN as a negative control.
(G) Luciferase reporter activities were assessed using MYC3×RE constructs in the presence of exogenous MYC in 293T cells (left) or when co-expressing TAL1 3×RE reporter constructs with TAL1 (right).
Data shown represent the means (±SD) of three biological replicates, **p < 0.01; significance was determined by unpaired two-tailed Student’s t test (F) or one-way ANOVA test followed by Tukey’s correction (C, D, and G). See also Figure S3.