Figure 5.
Proliferating CD4+ T Cells Contain Regulatory and Cytotoxic Cell States
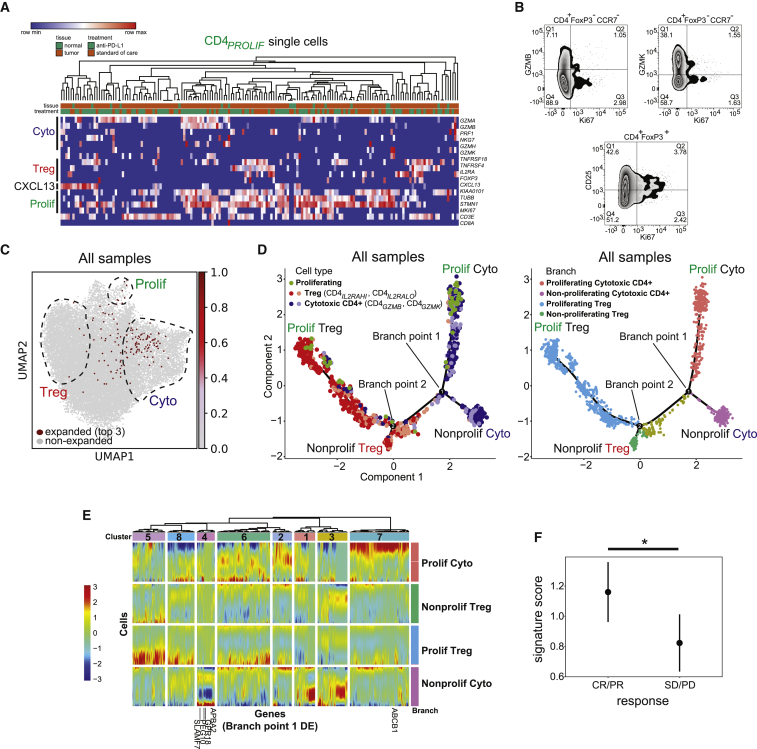
(A) Heatmap showing expression of select cytotoxic, regulatory, and proliferating marker genes (rows) for individual single cells (columns) within the CD4PROLIF cluster. Samples were hierarchically clustered. Log2-transformed expression of each gene was row scaled.
(B) Representative flow cytometry staining from a bladder tumor showing expression of CD25, GZMB, GZMK, and Ki67.
(C) Single cells expressing the top 3 most expanded clonotypes found in the CD4PROLIF T cell population are shown in red in the same UMAP space as in Figure 2A. The regions composed of proliferating, regulatory, and cytotoxic T cells are outlined and superimposed on the UMAP projection for visualization.
(D) Left panel: pseudotime trajectories derived from all tumors (N = 7 samples) and non-malignant samples (N = 6 samples). Cells with expanded TCRs from the proliferating (CD4PROLIF, green), regulatory (CD4IL2RAHI and CD4IL2RALO, shades of red), and cytotoxic (CD4GZMB and CD4GZMK, shades of purple) states were used for this analysis. Specific branches corresponding to proliferating cytotoxic cells (top right), non-proliferating cytotoxic cells (bottom right), proliferating regulatory cells (top left), and non-proliferating regulatory cells (bottom left) are labeled. Right panel: branches are color-coded according to the above proliferating or non-proliferating identities. Also labeled are branch points that discriminate proliferating and non-proliferating cytotoxic CD4+ T cells (branch point 1) and proliferating and non-proliferating regulatory T cells (branch point 2).
(E) Heatmap showing all differentially expressed genes (columns) between branches for branch point 1 across cells in the pseudotime analysis (rows). Cells are grouped by their proliferating or non-proliferating branch assignments, color-coded at the right of the heatmap and corresponding to colors in (D). Genes are grouped by color-coded clusters (1–8) shown at the top of the plot, which result from hierarchical clustering based on co-regulation in specific branches.
(F) Cytotoxic CD4+ T cell gene signature scores were plotted in clinical responders (complete response or partial response) versus non-responders (stable disease or progressive disease) from baseline metastatic biopsies from bladder cancer patients with inflamed tumors on the IMvigor210 clinical trial (N = 62 tumors). The signature score was obtained from the IMvigor210 bulk RNA-seq dataset for the cytotoxic CD4+ T cell-specific genes derived from non-proliferating (cluster 4) and proliferating (cluster 7) cytotoxic CD4+ clusters from the pseudotime analysis shown below the heatmap in (E). Median ± SEM is shown; ∗p = 0.037 by two-tailed t test.