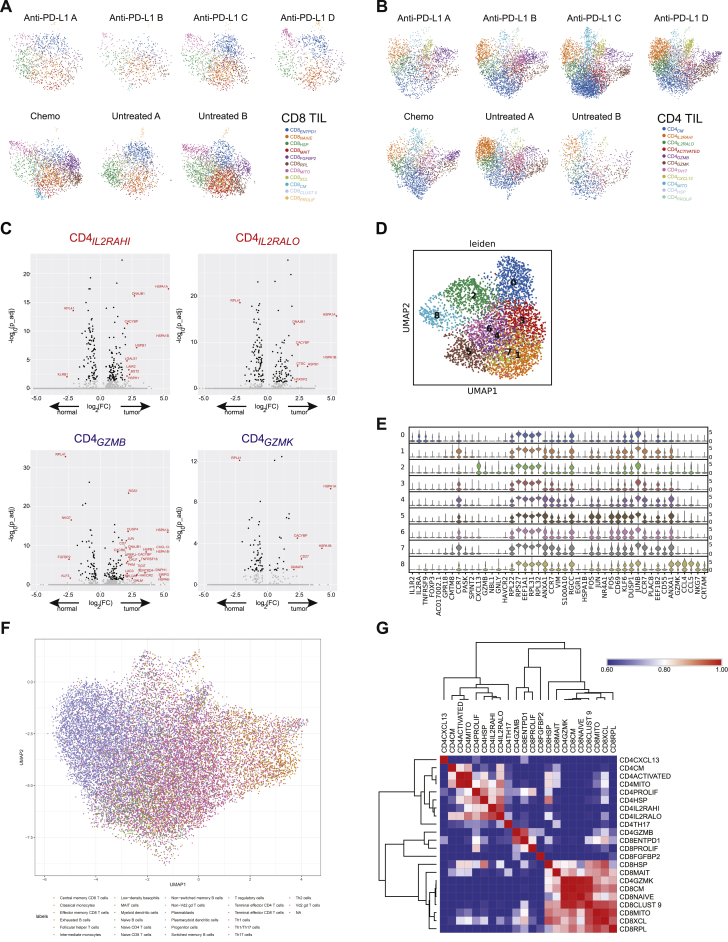
Figure S2.
Clustering, Differential Expression, Annotation, and Correlation Analysis of T Cell Transcriptional Phenotypes, Related to Figures 1, 2, 3, and 4
(A-B) UMAP plots showing cluster representation for CD8+ (A) and CD4+ (B) TIL from individual patients. (C) Volcano plots showing adjusted P values versus log2(FC) for differential testing of genes between tumor and non-malignant compartments for regulatory T cell populations (top, CD4IL2RAHI, CD4IL2RALO) and cytotoxic CD4+ populations (bottom, CD4GZMB, CD4GZMK). Genes whose expression is significantly different between compartments with Padj < 0.05 and |log2(FC) > 1.4| are shown in red. (D-E) Unbiased clustering of CD4+ T cells from tumor and adjacent non-malignant tissue from a single patient (anti-PD-L1 C). (D) UMAP plot showing individual cells coded by cluster or by tissue of origin. (E) Violin plot showing top 5 differentially expressed marker genes for each unbiased cluster. (F) Annotations of single CD4+ T cells from tumor and adjacent non-malignant tissue using SingleR. (G) Correlation matrix of all CD4+ and CD8+ populations from tissue (combined tumor and non-malignant tissues) based on expression of shared genes. Pearson correlation coefficient is shown. Populations were arranged based on hierarchical clustering using Euclidean distance metric.