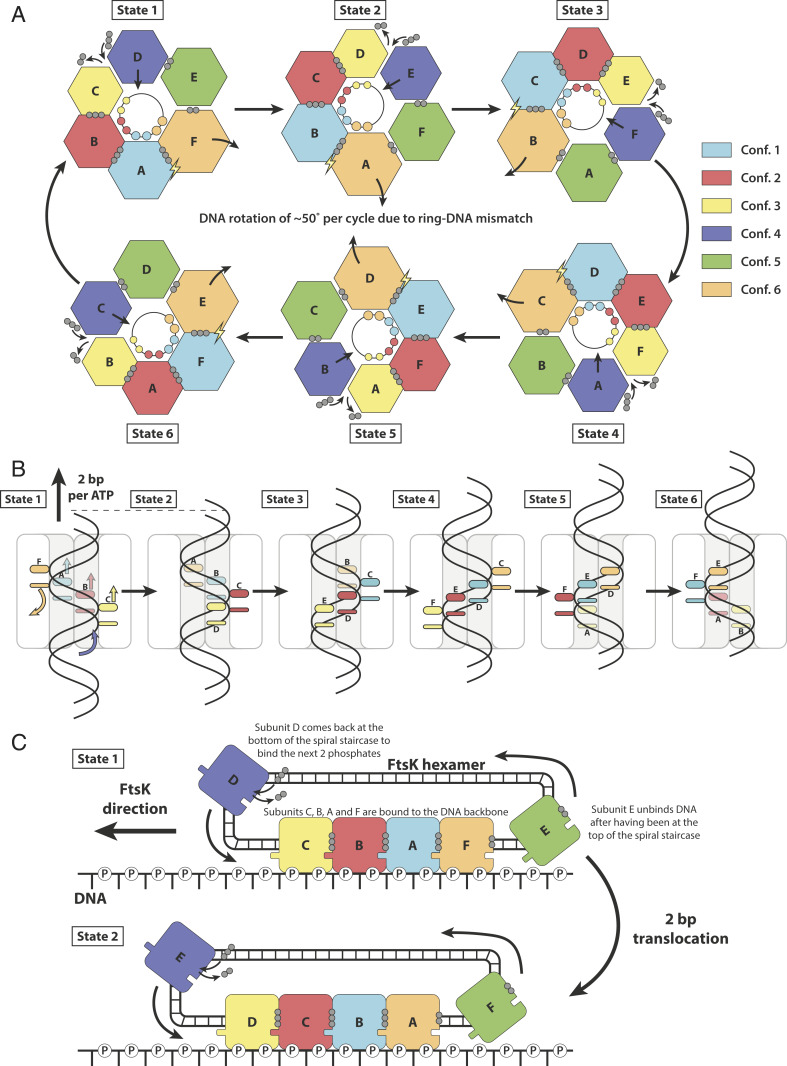
Fig. 4.
Model for double-stranded DNA translocation. (A) The six conformations rotate around the ring. As each subunit advances one-sixth further along the ATPase reaction cycle, it adopts the conformation of the subunit next to it. The conformations, but not the subunits, rotate clockwise around the ring (viewed from the α-ring). Because the DNA is helical, rotational adaption of the six conformations leads to 2-bp translocations at each step. Since DNA has 10.5 bp per turn, there needs to be a rotation of FtsK against the DNA of 51.4 ° per cycle of six hydrolyzed ATP and 12 bp translocated. Spheres: phosphates; lightning bolt: hydrolysis-competent state. (B) DNA translocation through the FtsKαβ pore. In each state, four subunits bind dsDNA through two loops, organized into two spiral staircases. (C) The translocation mechanism understood as circularized filament treadmilling. As in cytomotive filaments, the spiral staircase along the DNA backbone is extended at one end (conformation 3, yellow, binding DNA last), and shortened at the other end (conformation 6, orange, detaching next). Non–DNA-binding conformations 4 and 5 circularize the filament into a ring since they close the conformational wave, resetting conformation 6 into 3.