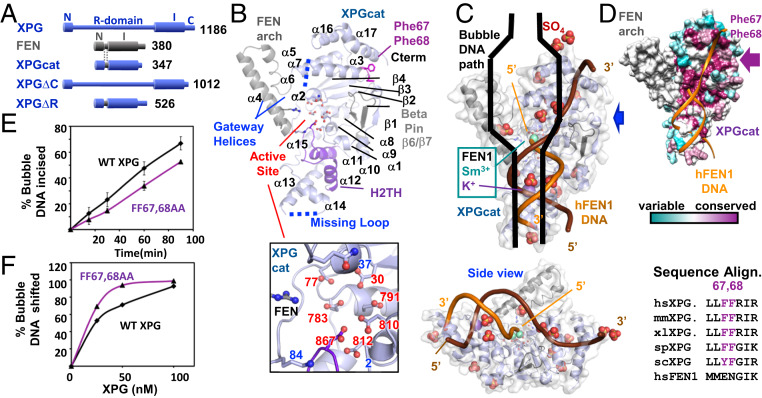
Fig. 1.
Conserved features in XPGcat structure. (A) Domain schematic of XPG, FEN, and XPG deletion mutants. In XPGcat and XPG∆R, XPG N and I catalytic regions have replaced the R-domain with the corresponding region in FEN. (B) XPGcat cartoon denoting FEN superfamily elements. Zoom-in view shows XPGcat active site carboxylates and gateway basic residues and Gln37. (C) Orthogonal views of overlay of FEN1:DNA (PDB ID code 3Q8K) onto XPGcat surface representation and SO4’s. Only DNA, K+, and catalytic metals from the FEN1:DNA complex are shown. Blue arrow gives side view perspective. Black cartoon of bubble DNA suggests that only half a bubble DNA would fit on the DNA binding face of XPGcat. (D) XPGcat surface representation colored by conservation. Sequence alignment shows that Phe67 and Phe68 are highly conserved residues in XPG family, but not in FEN1. (E) Incision activity of 15-nt bubble DNA (HP15T/C) by FF67,68AA was decreased relative to WT XPG. Single turnover assays done in triplicate with 33.3-nM flag-tagged protein and 3.3-nM DNA. Error bars show SD. Representative gel in SI Appendix, Fig. S1F. (F) FF67,68AA had higher affinity for 15-nt bubble DNA (5 nM) compared to WT XPG in EMSA. Representative gel in SI Appendix, Fig. S1G.