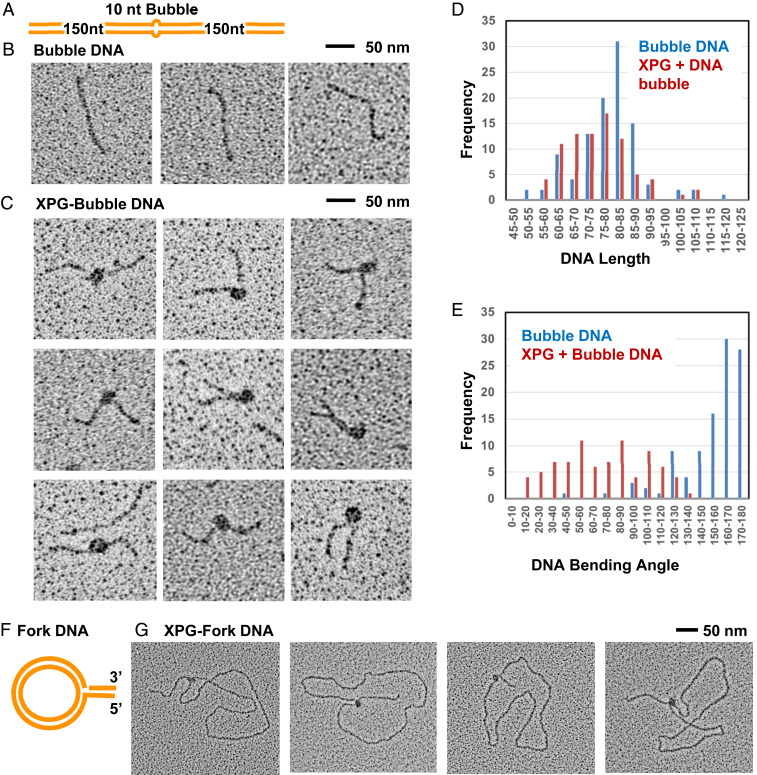
Fig. 4.
EM analysis shows direct interaction of XPG with bubble DNA and forks. EM analysis included tungsten rotary metal shadow casting following mounting of protein or DNA onto thin carbon foil substrates. (A) Schematic of the 10-nt bubble DNA EM substrate. (B) EM analysis showed bubble DNA alone was mostly straight. (C) XPG and bubble DNA with 5 mM Ca+2. A distinct protein particle of a size consistent with an XPG dimer was observed at the center of a large fraction of the DNA, but never at the end. Protein-bound DNA frequently had severe bends. (D) Histogram of measured XPG-free and XPG-bound DNA lengths. To determine if the DNA wrapped around XPG, the length of protein free DNA (blue), or DNA containing XPG bound as in C were measured. The two length distributions are similar arguing against DNA wrapping. (E) Histogram of XPG-free and XPG-bound DNA bending angles, determined from the EM images; “180” corresponds to a straight DNA, while a value of 0 corresponds to a DNA fully folded back on itself at the center. (F) Schematic of the single replication fork EM substrate. (G) The fork DNA was incubated with XPG with 5 mM Ca+2 and analyzed by EM. XPG protein particles, often oblong, were present only at the replication fork.