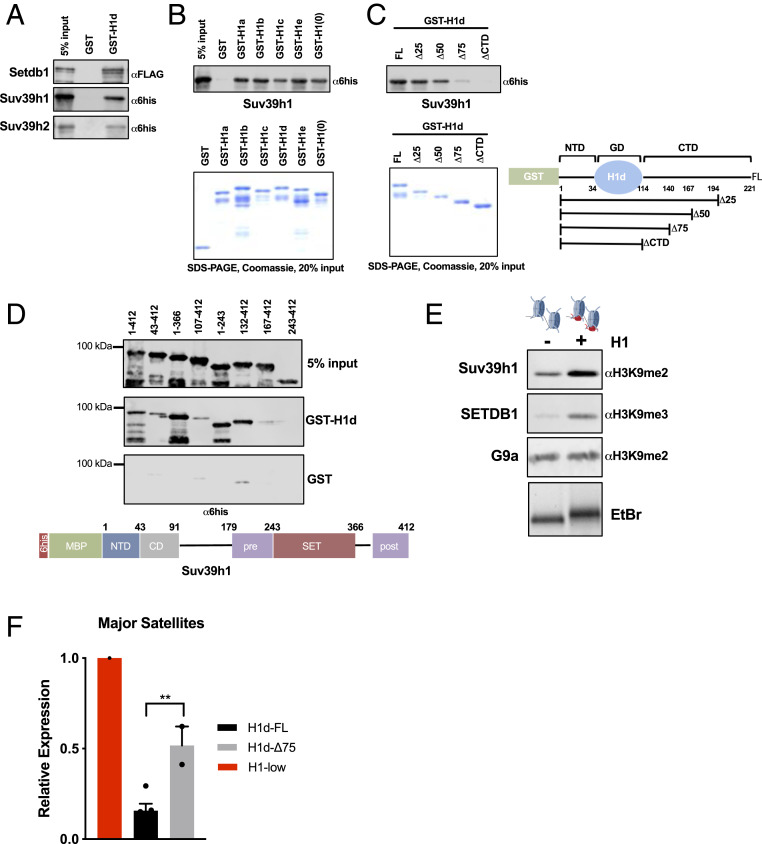
Fig. 3.
Linker histones interact with Suv39h1/h2 and SETDB1 and promote H3K9 methylation in vitro. (A) Interaction studies between H1 and H3K9 methyltransferases. Linker histone GST fusion proteins were purified, bound to glutathione Sepharose beads, and incubated with the indicated recombinant proteins tagged with either hexahistidine-maltose binding protein (6his-MBP) or Flag peptides. Bound proteins and 5% input control samples were analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) followed by immunoblotting for the indicated peptides. (B and C) GST pulldown assays as in A with the indicated proteins. Coomassie blue-stained SDS/PAGE gel of purified proteins showing equal loading (Lower). (C, Right) Schematic representation of GST-H1d polypeptides used in interaction assays. Numbers indicate amino acid residues. (D) GST pulldown assays as in A with the indicated polypeptide fragments of Suv39h1. After incubation with GST or GST-H1d, pulldown products and 5% input controls were analyzed with a hexahistidine(6his)-specific antibody. A schematic illustration of Suv39h1 domain structure is shown below. (E) In vitro HMT assays with reconstituted chromatin in the presence or absence of H1. Chromatin was reconstituted in vitro using a DNA template bearing two repeats of the synthetic “601” nucleosome positioning sequence and recombinant histone octamers in either the presence or absence of H1. H1 incorporation was verified by nondenaturing agarose gel electrophoresis followed by ethidium bromide (EtBr) staining, demonstrating slower migration of H1-containing dinucleosomes (Lower). Dinucleosomes were incubated with the indicated enzymes (Suv39h1, 100 nM; SETDB1, 50 nM; G9a, 20 nM) under conditions described in Materials and Methods. Enzymatic activity was detected by immunoblotting for the indicated histone modifications. (F) RT-qPCR of major satellite transcripts. Expression of major satellites was quantified in H1-low lines in which either full-length H1d (H1d-FL) or H1d lacking 75% of the CTD (H1d-Δ75) bearing an N-terminal 3xFlag tag were reintroduced via stable transfection. Expression (relative to the parental line) was calculated using the ΔΔCt method normalized to Gapdh mRNA. n = 2 or 3 independent clones per condition.