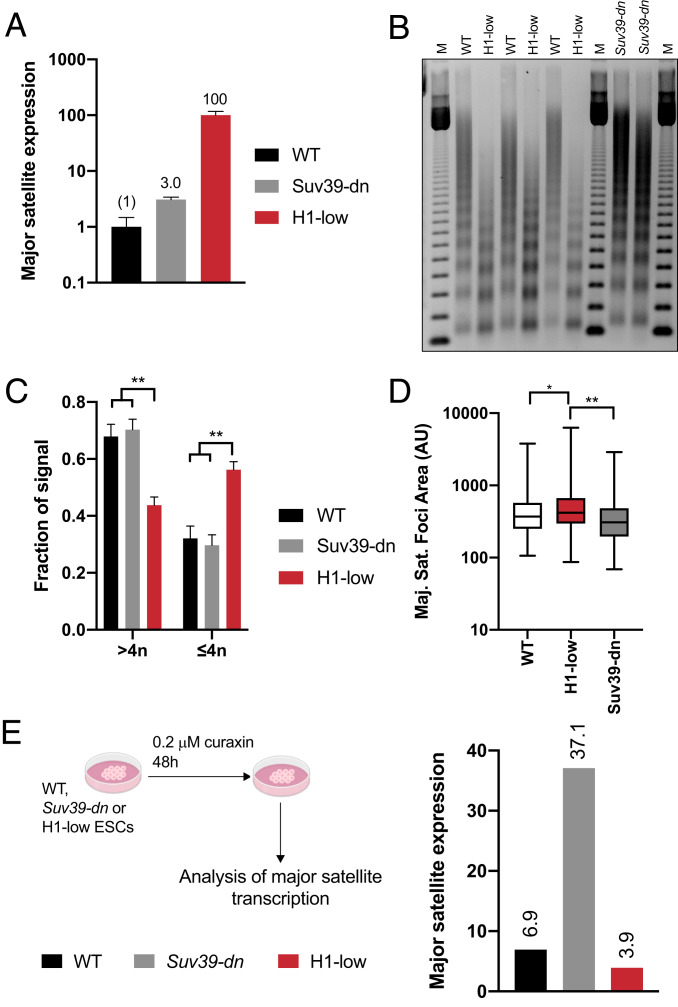
Fig. 4.
H1-mediated chromatin compaction is the dominant mechanism of major satellite repression. (A) Expression of major satellite transcripts in WT, Suv39-dn, and H1-low ES cells. Major satellite transcripts in WT, Suv39-dn, and H1-low ES cells were measured by RT-qPCR and normalized to Gapdh mRNA using the ΔΔCt method. Numbers above indicate expression relative to WT. Data are from three technical replicates of the indicated cell line. Similar results were obtained with several lines of the same genotype (see Fig. 2A). (B) Nucleosome spacing in WT, Suv39-dn, and H1-low ES cells. Nuclei were subjected to limited digestion with micrococcal nuclease and DNA was purified and analyzed by nondenaturing agarose gel electrophoresis, followed by ethidium bromide staining. (C) Analysis of chromatin condensation in WT, Suv39-dn, and H1-low ES cells. The fraction of signal arising from DNA fragments in B corresponding to tetranucleosomes and below (≤4n) and pentanucleosomes and above (>4n) was quantified and normalized to the total signal in each lane using ImageJ. n = 2 or 3 clones per genotype; error bars represent SD. (D) Area of major satellite foci in WT, Suv39-dn, and H1-low ESCs. At 24 h after transfection with TALEN specific to major satellite DNA fused to mClover, cells were sorted, applied to coverslips, and fixed. The area of major satellite foci was determined using ImageJ. WT, n = 180; Suv39-dn, n = 191; H1-low, n = 279. *P ≤ 0.05; **P < 0.01. Unpaired t test with Welch’s correction was used to test statistical significance. (E) Expression of major satellites in dimethyl sulfoxide (DMSO)- and curaxin-treated cells. Major satellite transcripts in WT, Suv39-dn, and H1-low ES cells treated with 0.2 µM curaxin or DMSO for 48 h were measured by RT-qPCR as in A. Data are shown as relative to DMSO control at 24 h.