Figure 2.
GIV Is Required for Exo70 Localization and Polarized Exocytosis
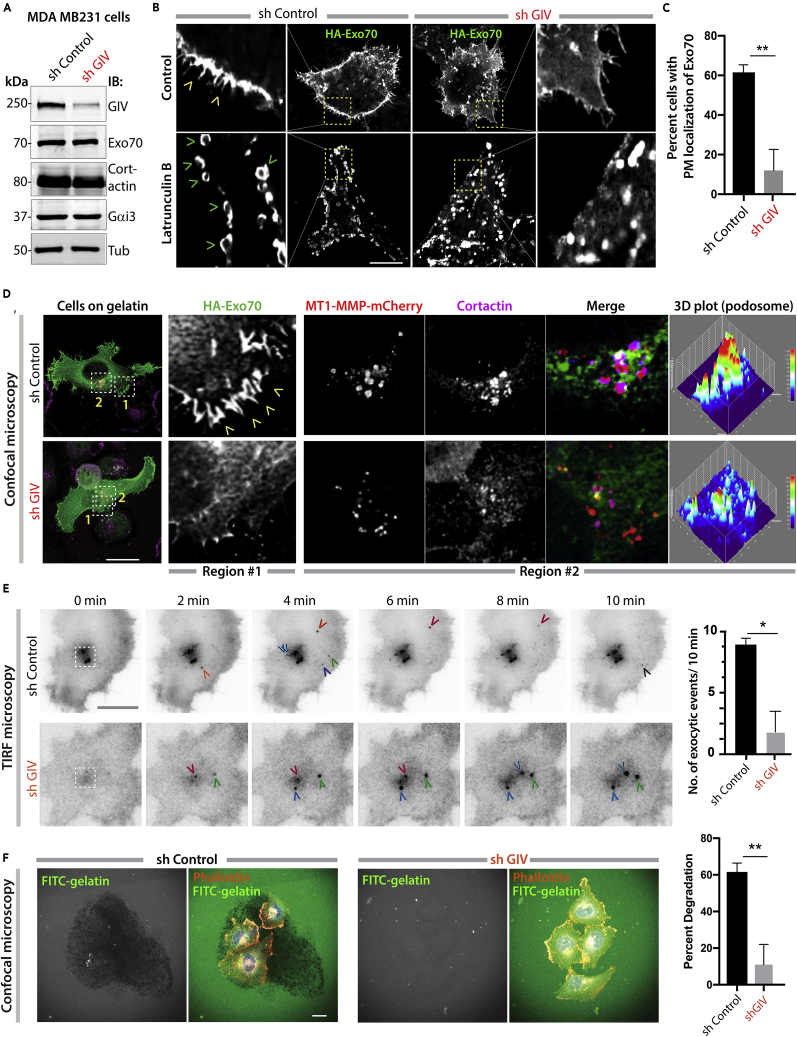
(A) Immunoblots of lysates of control (sh Control) and GIV-depleted (sh GIV) MDA MB231 cells.
(B) Control (sh Control) and GIV-depleted (sh GIV) MDA MB231 cells expressing HA-Exo70 were treated (bottom) or not (top) with 25 μM latrunculin B for 24 h before fixation with PFA. Fixed cells were stained for HA and analyzed by confocal microscopy. Representative deconvolved images are shown. Scale bar, 10 μm. Arrowheads, PM localization.
(C) Bar graphs show the % cells with PM localization of Exo70-HA. Error bars represent standard error of mean; ± SEM; n = ∼ 30 cells/assay, 3 independent assays. ∗∗p value <0.01.
(D) Cells in (B) exogenously expressing HA-Exo70 (green) and MT1-MMP-mCherry (red) were plated on gelatin-coated coverslips and fixed and stained with the invadosome marker cortactin (magenta). Representative deconvolved images are shown. Boxed area with grey interrupted line represents cell's center, likely invadosomes. Colored arrowheads track single exocytic events. Scale bar, 10 μm. Insets were analyzed by rendering 3D surface plots using ImageJ. Cortactin/MT1-MMP colocalization was determined by Pearson's correlation coefficient reaching a value of 0.71 ± 0.05 and 0.15 ± 0.04 in control and GIV-depleted cells, respectively, from four different experiments.
(E) Cells in (B) exogenously expressing MT1-MMP-pHLuorin were plated on a layer of unlabeled gelatin and imaged by TIRF microscopy. Left: Still frames from a 10-min-long movie (Videos S1 and S2). Each colored arrowhead marks one successfully exocytosed vesicle. Scale bar, 2.5 μm. Right: Bar graph shows the number of exocytic events that were encountered during the 10-min-long movie in (D). N = ∼ 5–10 cells/assay. Error bars represent standard error of mean; ± SEM. ∗p value <0.05.
(F) Cells in (B) were plated for 5 h on fluorescein isothiocyanate (FITC)-conjugated cross-linked gelatin (green) and then fixed and stained for F-actin (Phalloidin; red). (Left) Representative images are shown. Scale bar, 10 μm. (Right) Bar graphs display the % of cells that showed degradation of gelatin. N = ∼100–200 cells/experiment × 3. Error bars represent standard error of mean; ± SEM. ∗∗p value = 0.001.