Figure 8.
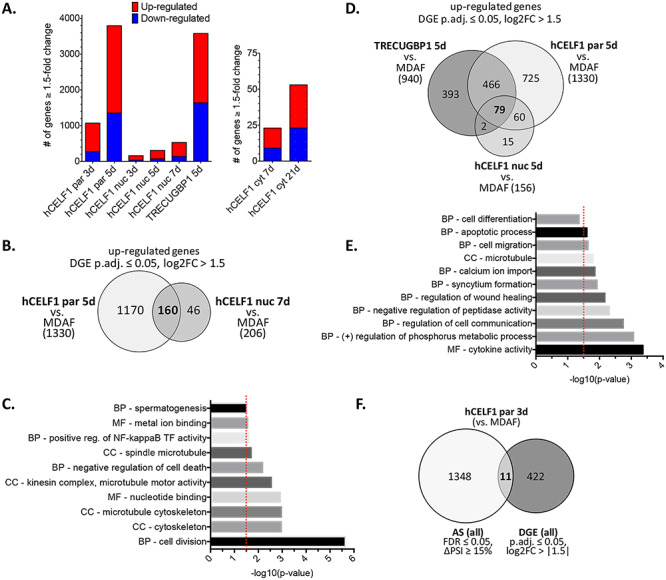
CELF1 overexpression results in widespread DGE in adult skeletal muscle. RNA-sequencing data analyzed for DGE. Animals with comparable mCherry-CELF1 protein expression and similar minimal histological defects were chosen for sequencing. (A) Differentially expressed genes displaying ≥1.5-fold change (P.adj ≤ 0.05) as compared to MDAF negative controls. (B) Overlap of DGE as compared to MDAF controls in the hCELF1par and hCELF1nuc lines induced for 5 or 7 days, respectively. (C) GO analysis of overlapping DGE between hCELF1par 5 day and hCELF1nuc 7-day induction. (D) Overlap of DGE as compared to MDAF controls in the hCELF1par, hCELF1nuc and TRECUGBP1 lines induced for 5 days. (E) GO analysis of overlapping DGE hCELF1par, hCELF1nuc and TRECUGBP1 lines induced for 5 days. (F) Genes undergoing AS transitions (∆PSI ≥ 15%) were overlapped with differentially expressed genes (log2-fold change >1.5) in hCELF1par induced for 3 days (as compared to MDAF negative controls). P, Fisher’s exact test. Red line = P-value of ≤0.032. CC, cellular compartment; BP, biological process; MF, molecular function.
