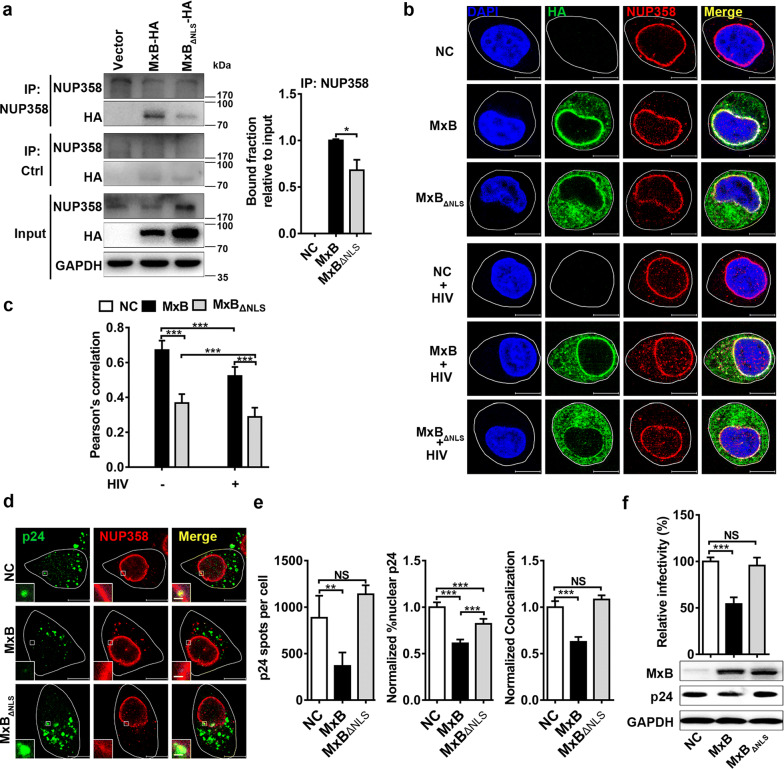
Fig. 1.
MxB binds to NUP358 and disrupts NUP358 association with HIV-1 viral cores. a The interaction between NUP358 and MxB was identified by co-immunoprecipitation analysis in 293T cells. Similar results were obtained in 3 independent experiments, and the standard deviation for the bound fraction relative to input was shown. b HeLa cells were transfected with plasmid expressing wild-type MxB-HA or MxBΔNLS-HA protein. 48 h after transfection, cells were infected with VSV-G pseudotyped HIV-1 luciferase reporter virus or not. Cells were fixed 6 h following infection and stained for MxB (green), NUP358 (red) using HA- or NUP358-specific antibodies, respectively. Cell nuclei were stained by DAPI (blue). Scale bar, 10 μm. c Pearson correlation coefficient values for colocalization of MxB with NUP358 were analysed by image analysis software in about 40 cells. Data was representative of 3 independent experiments. d HeLa cells were transfected with plasmid expressing wild-type MxB-HA or MxBΔNLS-HA protein. 48 h after transfection, cells were infected with VSV-G pseudotyped HIV-1 luciferase reporter virus. Cells were fixed 6 h post infection and stained for HIV-1 capsid protein p24 (green), NUP358 (red) and DAPI (blue) for cell nuclei. Scale bar, 10 μm. e P24 spots, the percentage of the p24 signals detected in the nucleus and the percent p24 colocalizing with NUP358 in HeLa cells was counted in about 40 cells. Data were representative of 3 independent experiments. f HeLa cells were transfected with plasmid expressing wild-type MxB-HA or MxBΔNLS-HA protein. 48 h after transfection, cells were infected with VSV-G pseudotyped HIV-1 luciferase reporter virus. The infectivity was determined 48 h post infection. Results were a summary of 3 independent experiments. Statistical significance: NS: not significant, *p < 0.05, **p < 0.01 and ***p < 0.001