Figure 1.
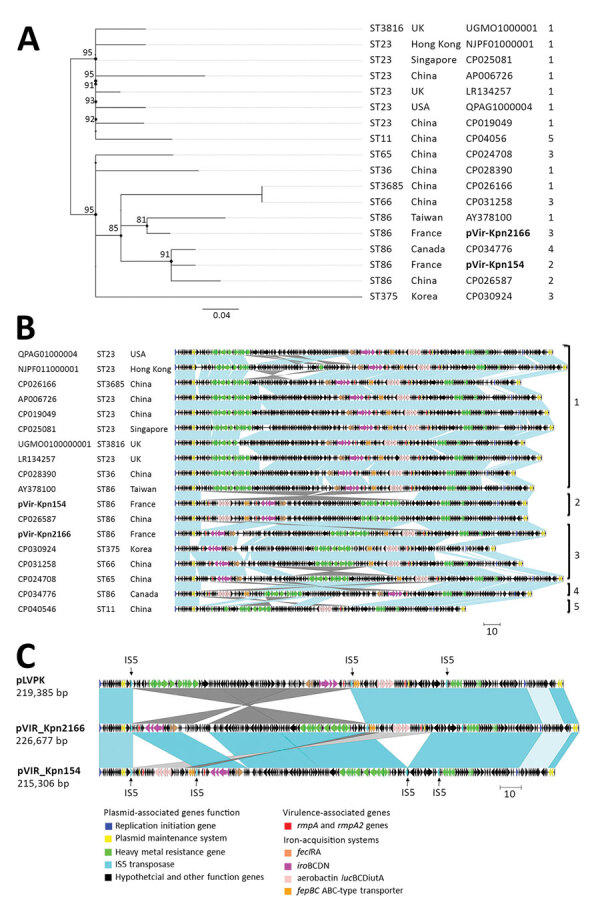
Comparison of pVIR-Kpn2166 and pVIR-Kpn154 Klebsiella pneumoniae isolates from 2 patients in France (bold) with 16 hypervirulent K. pneumoniae virulence plasmids recovered from the PATRIC database (http://www.patricbrc.org). A) Single-nucleotide polymorphism–based phylogenetic tree built by RaxML from an alignment generated by Burrows-Wheeler Aligner and filtered to remove recombination using Gubbins as previously described (9). The ST and the geographic origin of bacterial hosts are shown. Scale bar indicates mean number of nucleotide substitutions per site. B) Synteny analysis of hypervirulent K. pneumoniae virulence plasmids based on data from blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Virulence-based synteny groups are indicated and the operons encoding the virulence factors, virulence synteny groups of plasmids and the ST and the geographic origin of bacterial hosts. Scale bar indicates kbp. C) Comparison of the details of rearrangements observed in pVIR-Kpn2166 and pVIR-Kpn154 and in pLVPK. Scale bar indicates kbp. ST, sequence type.
