Figure 4.
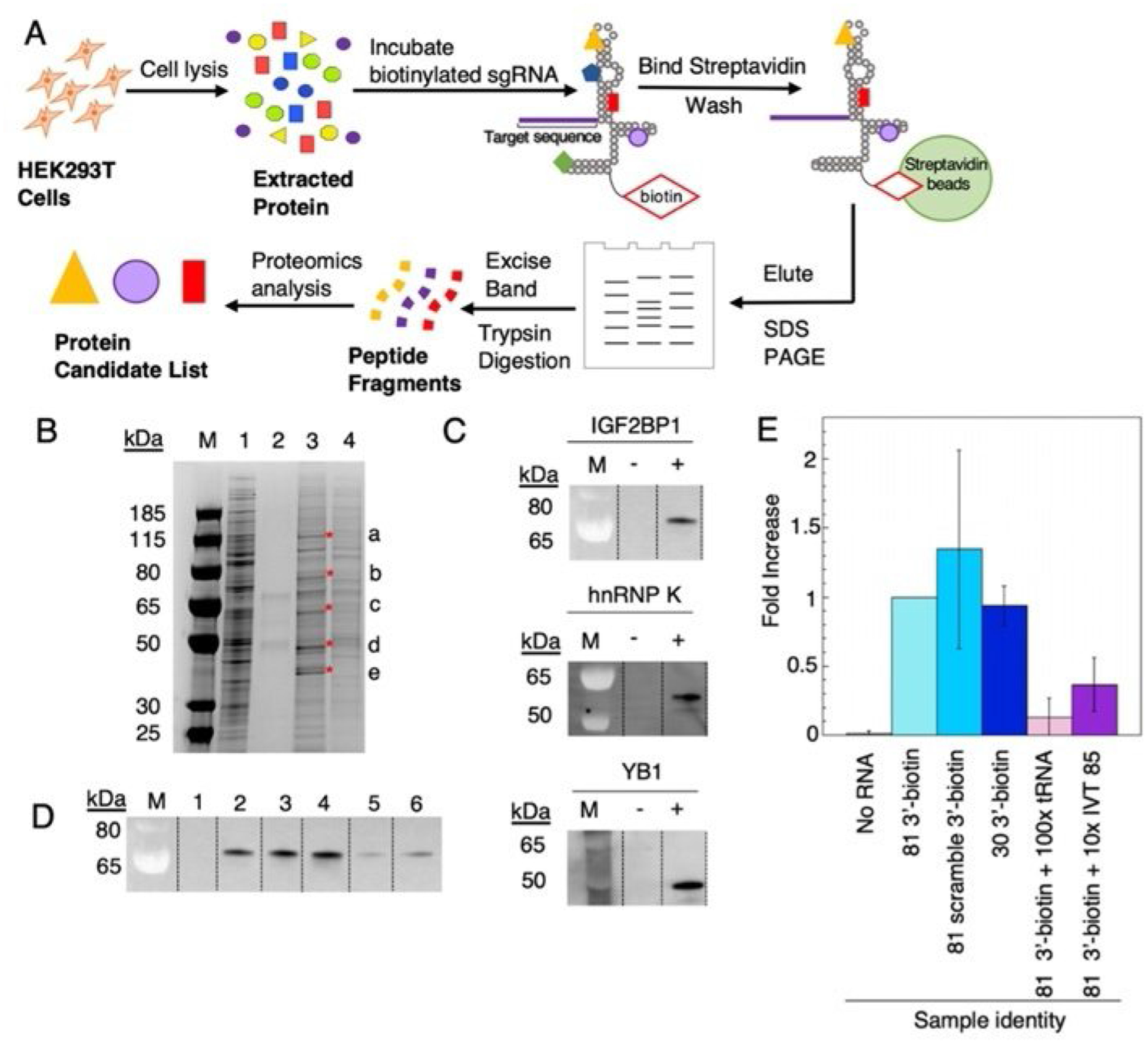
Identification of endogenous human sgRNA-binding proteins. A) Workflow scheme for identification of candidate endogenous human sgRNA binding proteins from HEK 293T cell lysate. B) SDS-PAGE gel stained with Imperial protein stain. Lanes: M=ladder, 1=5 μL of HEK 293T cell lysate, 2=no RNA control, 3= 81 3′-biotin pulldown, 4=81 3′-biotin +100-fold excess tRNA. Excised bands are indicated by a, b, c, d, e and red asterisks. C) Western blot verification of IGF2BP1, hnRNP K, YB1 present in elution from streptavidin beads. D) IGF2BP1 binding to sgRNA variants. Lanes: 1=no 81 3′-biotin, 2=81 3′-biotin, 3=81 3′-biotin with scrambled S1 target sequence (Table S1), 4= 5′P-30 nt-3′-biotin fragment, 5=81 3′-biotin+100-fold excess tRNA, 6=81 3′-biotin+ tenfold excess of IVT 85. Data in plot represents triplicate. E) Fold increase vs. RNA identity plotted. Plotted values correspond to averages±standard deviation (n=3).
