Figure 4. (p)ppGpp binds the conserved XPRT active site, and a flexible loop covering the pocket differentiates between pppGpp and ppGpp/pGpp.
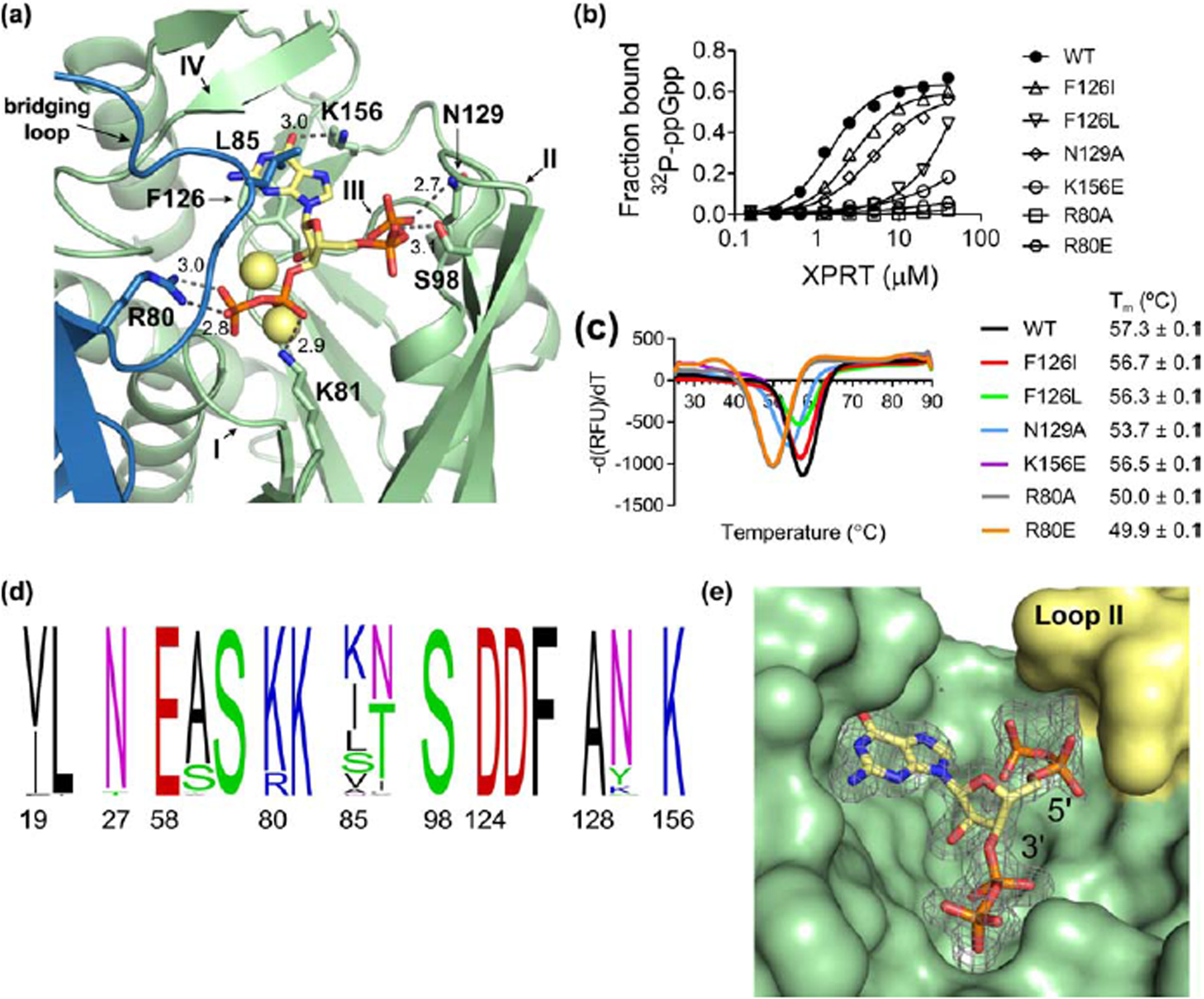
(a) Select residues involved in the ppGpp-XPRT interaction. Hydrogen bonding shown as dotted lines and is measured in Å. The blue protein is the adjoining monomer in the structure. Yellow spheres are Na+ ions. (b) DRaCALA of B. subtilis XPRT active site variants shows that 32P-ppGpp binding is weakened by altering these residues. Points are mean of triplicate for all but F126L (duplicate). Error bars represent SEM for triplicate and range for duplicate. Error bars may be smaller than the height of the symbols. (c) Derivative curves from differential scanning fluorimetry of XPRT variants. Curves show the mean of triplicate reactions. Melting temperature (Tm) is the mean of three replicates ± SEM. (d) Frequency logo of the ppGpp-binding residues from 62 bacterial XPRTs. Numbering is according to B. subtilis XPRT. Residues are colored according to their class. Logo generated using WebLogo (UC Berkeley). (e) Surface view of the ppGpp binding pocket on XPRT. Omit difference density for ppGpp is shown contoured at 2.5 σ. The compression around the 5’-phosphates caused by loop II (yellow) may be responsible for weaker interaction with pppGpp than ppGpp/pGpp.
