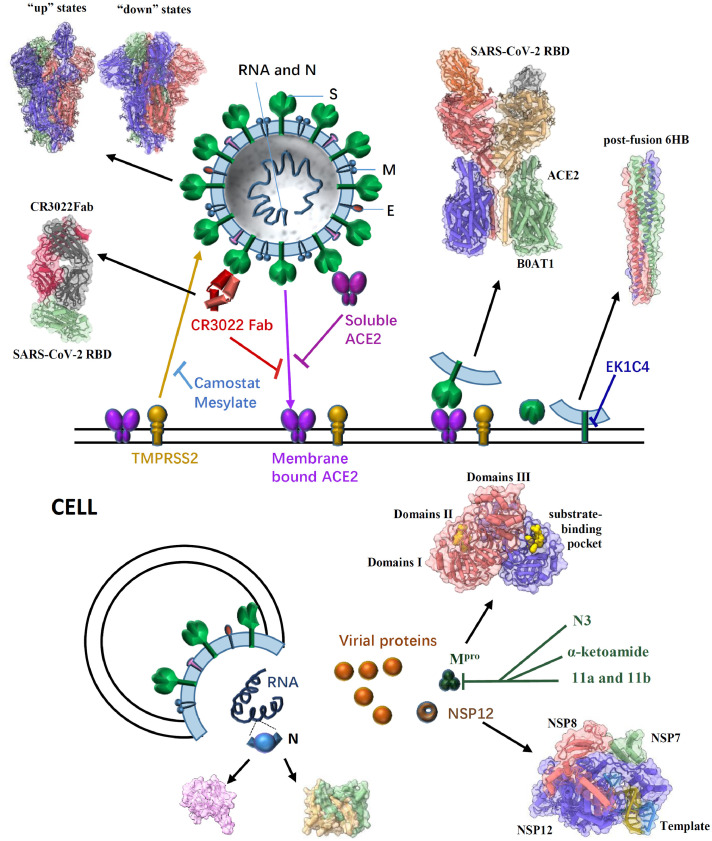
Fig. 2.
General structure of a SARS-CoV-2 virion, the pre- and post-fusion states of the SARS-CoV-2 S glycoprotein, and some effective inhibitors targeting different proteins to impair SARS-CoV-2 infection. SARS-CoV-2 enters host cells mainly through the trimeric S glycoprotein (PDB ID: 6VSB, 6VYB/6VXX) containing RBD which binds to the human receptor ACE2 (PDB ID: 6M17). A previously characterized SARS-CoV monoclonal antibody CR3022 could bind to the SARS-CoV-2 RBD (PDB ID: 6W41); the epitope can potentially confer in vivo protection. The recombinant soluble human ACE2 can bind to RBD of the S protein to block the pre-fusion stage of SARS-CoV-2 infection. The TMPRSS2 for S protein priming can be inhibited by camostat mesylate, which in turn would block SARS-CoV-2 infection. After receptor binding of the S1 subunit to support membrane fusion and viral infection, the S2 subunit is exposed and undergoes a conformational change from a pre-fusion to a post-fusion state (PDB ID: 6LXT). The inhibitor EK1C4 targets the S2 to inhibit viral infection. In cells, SARS-CoV-2 RNA translates to different proteins, such as NSP12 (PDB ID: 7BV1), N protein (PDB ID: 6M3M & 6WZO) and Mpro (PDB ID: 6Y2G). Several compounds (N3, α-ketoamide,11a and 11b) have been identified to exhibit high inhibitory activity against SARS-CoV-2 Mpro activity and viral infection.