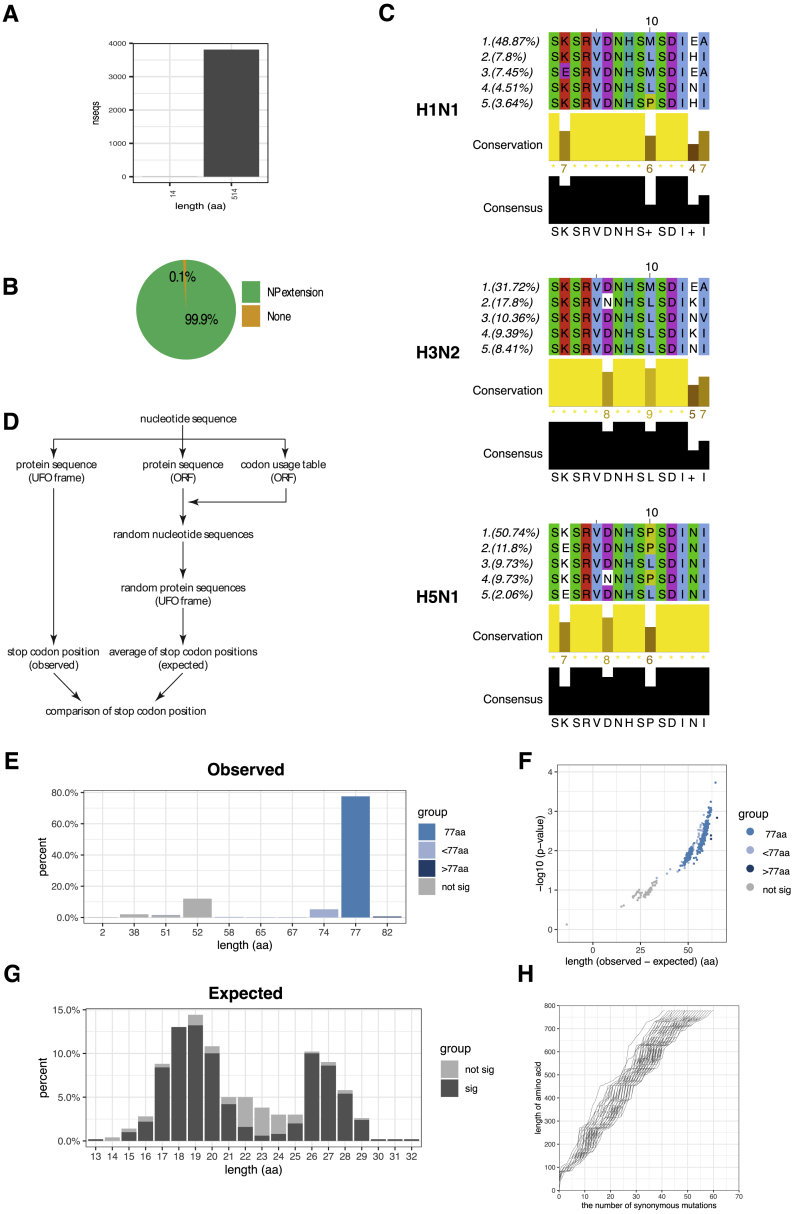
Figure S5.
uvORFs Are Conserved, Related to Figure 5
(A) Bar plot showing the number of unique NP sequences that give rise to the full length, extended NP protein of ∼514aa, or those that result in truncated (non-extended) uvORFs.
(B) Percentages of unique NP sequences that preserve the propensity to code for NP-extension.
(C) Top five most common NP extension protein sequences in three types of influenza A strains, H1N1, H3N2 and H5N1.
(D) Schematic showing the model used to calculate the expected versus observed PB1-UFO sequence lengths.
(E) Density plot of predicted length of H3N2 PB1-UFO protein sequences. Sequences predicted to generate a protein of 77aa are shown in medium blue, shorter than 77aa in light blue, and those longer than 77aa are in dark blue. Sequences predicted not to generate PB1-UFO protein are shown in gray.
(F) P value distribution/volcano plot of H3N2 PB1-UFO protein sequence length. Each dot represents the difference between observed length and expected length of each individual sequence.
(G) Density plot showing the distribution of expected lengths of H3N2 PB1-UFO proteins, based on random codon-shuffled sequences.
(H) Line plot showing the number of synonymous mutations in frame of WT H3N2 PB1 (x axis) that are required to generate stop codons in frame of H3N2 PB1-UFO (y axis).