Figure 1.
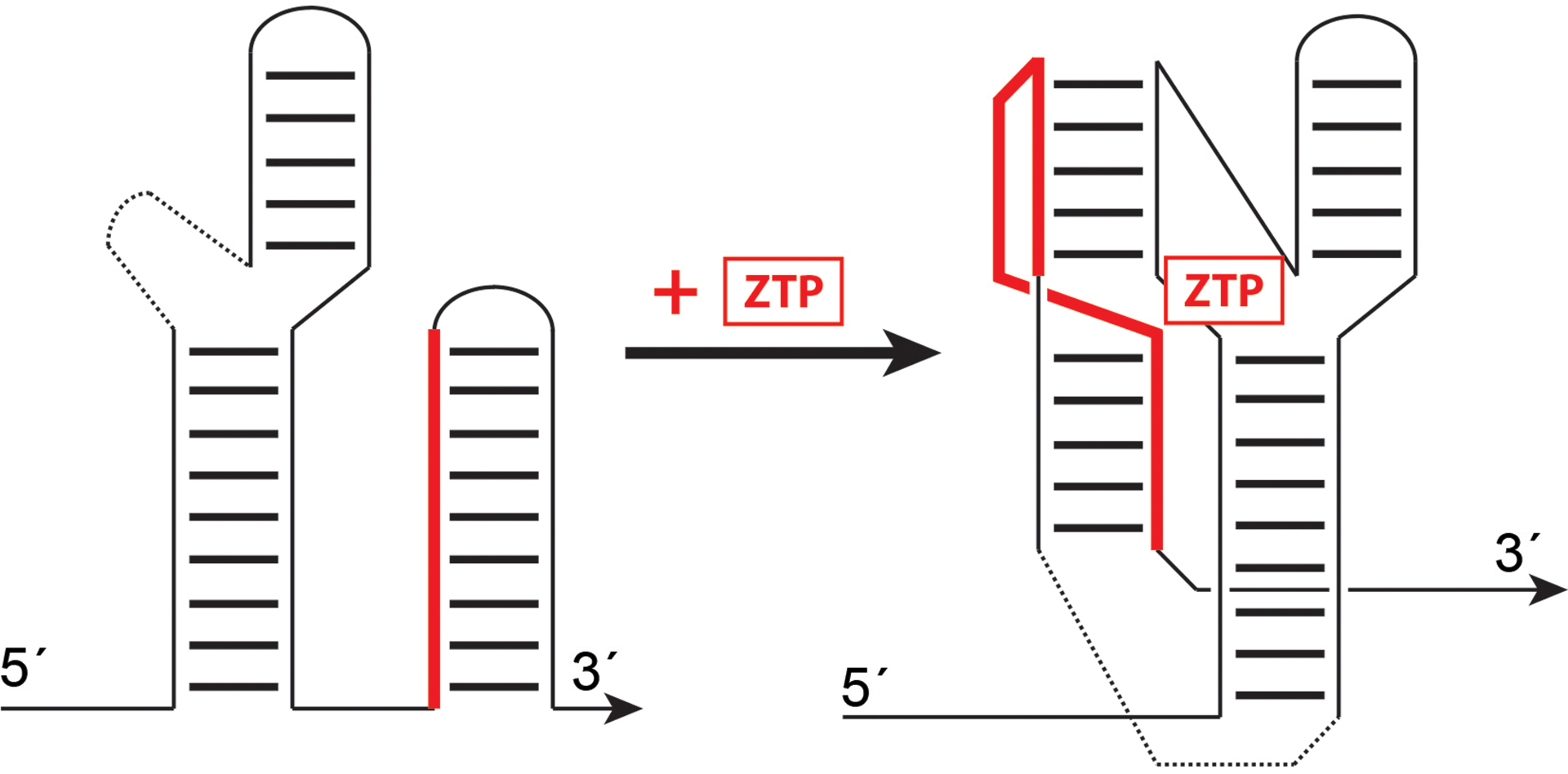
Found primarily in the 5´ untranslated regions of bacterial mRNAs, riboswitches alter gene expression in response to direct and specific small molecule binding. As exemplified by the ZTP riboswitch, the RNA exists in one conformational state in which a switch sequence (solid red) pairs to form a stable hairpin linked to transcription or translation regulation of a downstream 3´ open reading frame (not shown). In the presence of ZTP, an alternative pairing of the switch sequence is favored, resulting in an alternative secondary structure that binds ZTP and disfavors downstream hairpin formation.
