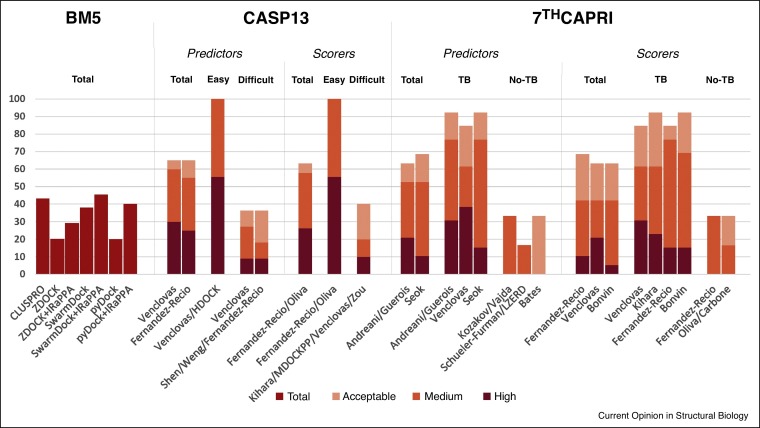
Figure 1.
Predictive success rates of state-of-the-art docking approaches on different benchmark sets. ClusPro performance on Protein–Protein Docking Benchmark 5.0 (BM5) is taken from Ref. [20•]. Performance of other docking methods on BM5 is taken from Ref. [26]. The rest of results are taken from CASP13-CAPRI and 7th CAPRI blind experiments. Sampling and scoring strategies included, but were not limited to: FFT-based sampling (ZDOCK, FTDock, ClusPro, Weng, Kozakov/Vajda, Shueler-Furman, Venclovas, pyDock, Fernandez-Recio, HDOCK, MDockPP, Zou, Shen, Seok), geometric hashing (Kihara, LZerD), particle swarm optimization (Bates), NMA-based sampling (Shen, Bates), information-driven sampling (Bonvin), energy-based scoring (pyDock, Fernandez-Recio), machine learning-based scoring (IRaPPA, Shen), statistical potentials (Kihara, MDockPP, Zou), evolutionary-based scoring (Andreani/Guerois), Voronoi-based scoring (Venclovas), shape-based scoring (HDOCK), docking-based contact consensus and residue propensities (Oliva, Carbone), and flexible refinement (Shueler-Furman, Seok).