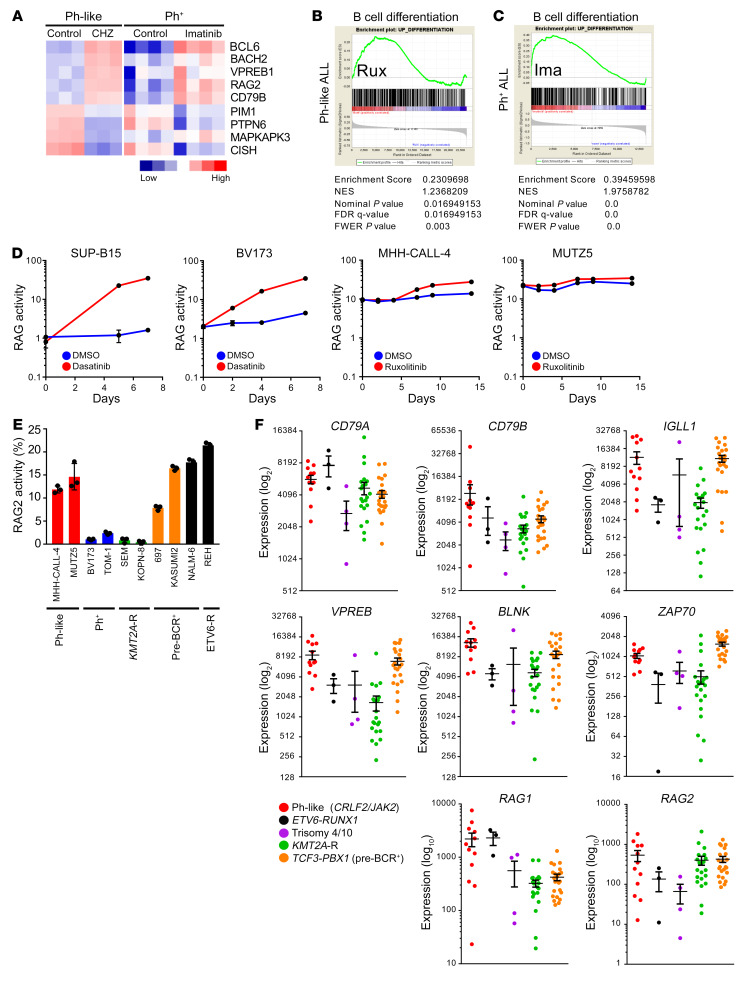
Figure 4. JAK inhibition induces partial differentiation of Ph-like ALL cells.
(A) Meta-analysis of gene expression data comparing MHH-CALL-4 cells treated with the type 2 JAK2 inhibitor CHZ868 with Ph+ ALL cells (BV173, NALM-1, SUP-B15, and TOM-1) treated with imatinib (12, 38). (B and C) Gene set enrichment analysis (GSEA) plots demonstrate enrichment of B cell differentiation gene sets in MHH-CALL-4 cells treated with ruxolitinib 1 μM for 12 hours (n = 3) (B) and Ph+ ALL treated with imatinib (38) (C); statistical analysis is shown. (D) Ph+ ALL cell lines and Ph-like ALL cell lines were lentivirally transduced with a RAG enzyme activity reporter construct. Ph+ and Ph-like ALL cells were treated with 100 nM dasatinib and 1 μM ruxolitinib, respectively. RAG enzyme activity was measured at the indicated times (n = 3 independent experiments). (E) Steady-state RAG enzyme activity was measured via flow cytometry in B-ALL cell lines (n = 3 independent experiments). (F) Supervised analysis of publicly available gene expression data from children with CRLF2-R/JAK2-mutant Ph-like ALL (n = 12), ETV6-RUNX1 ALL (n = 3), hyperdiploid ALL with trisomy 4 and 10 (n = 4), KMT2A-R ALL (n = 21), and TCF3-PBX1 ALL (n = 23) from the National Cancer Institute TARGET database. Data are represented as individual values with mean ± SEM bars.