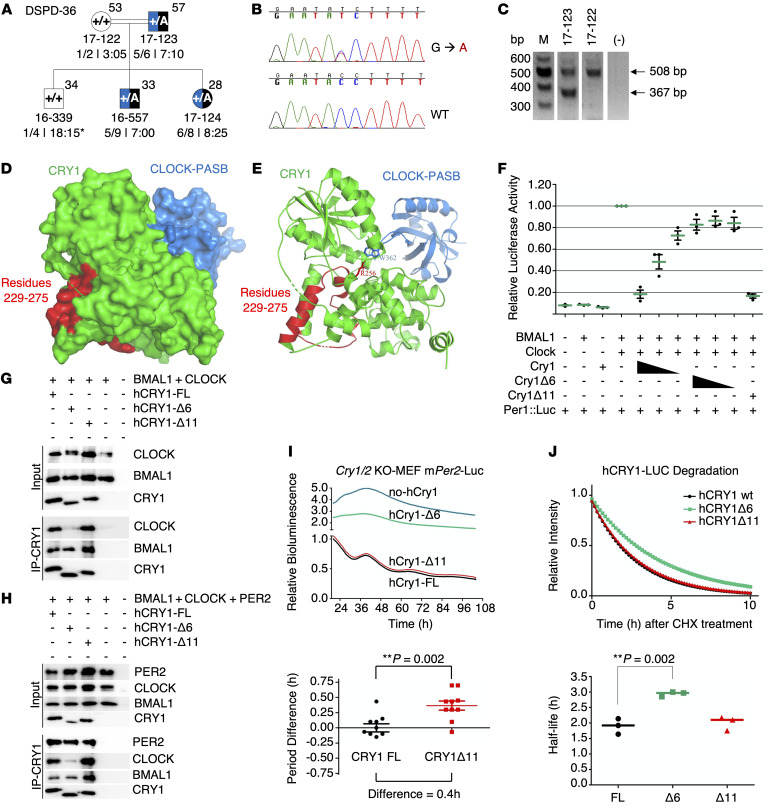
Figure 8. Phenotype-genotype characterization of family DSPD-36 and functional characterization of CRY1Δ6.
(A–C) Phenotype-genotype characterization of family DSPD-36. (A) The family DSPD-36 was assessed as described in the legend to Figure 3. (B and C) c.825+1G>A causes skipping of 141-bp exon 6, leading to an in-frame deletion of 47 residues in the middle of the PHR and clock-binding domains of the CRY1 protein. M, DNA marker. (D–J) Functional characterization of CRY1Δ6. (D) Docking analysis of CRY1 and the CLOCK PAS-B domain. (E) The critical amino acid residue of CRY1 (R256) interacts with CLOCK PAS-B (W362). (F) Analysis of the effect of FL WT CRY1, CRY1Δ6, and CRY1Δ11 on CLOCK/BMAL1-driven transcription with the Per1:Luc assay. An E-box–driven luciferase reporter plasmid Per1:Luc was coexpressed in HEK293T cells along with plasmids consisting of CLOCK and BMAL1 cDNAs and decreasing amounts of WT CRY1 or CRY1Δ6. Data represent the mean induction of bioluminescence over basal levels from duplicate transfections 48 hours after transfection. Error bars represent the SD from at least 3 biological experiments. Co-IP assay with (G) CLOCK, BMAL1, (H) PER2, CLOCK, BMAL1, and human CRY1s (hCRY1). Blots in G and H are representative of at least 3 independent experiments. (I) Rescue assays were performed with human CRY1s with at least 4 biological replicates. Samples were normalized to the initial luminescence signal. The graph below indicates the period differences, with the whiskers representing the mean ± SEM values. n = 5 per group. Data were pooled from 2 independent experiments. **P = 0.002, by unpaired t test. mPer2-Luc, mouse Per2-Luc. (J) Degradation assay with human CRY1::Luc and its variants. The graph below was generated by fitting a 1-phase decay curve to the data, which indicate the half-life, with the horizontal line representing the mean. n = 3 per condition, pooled from 3 independent experiments. **P = 0.002, by unpaired t test.