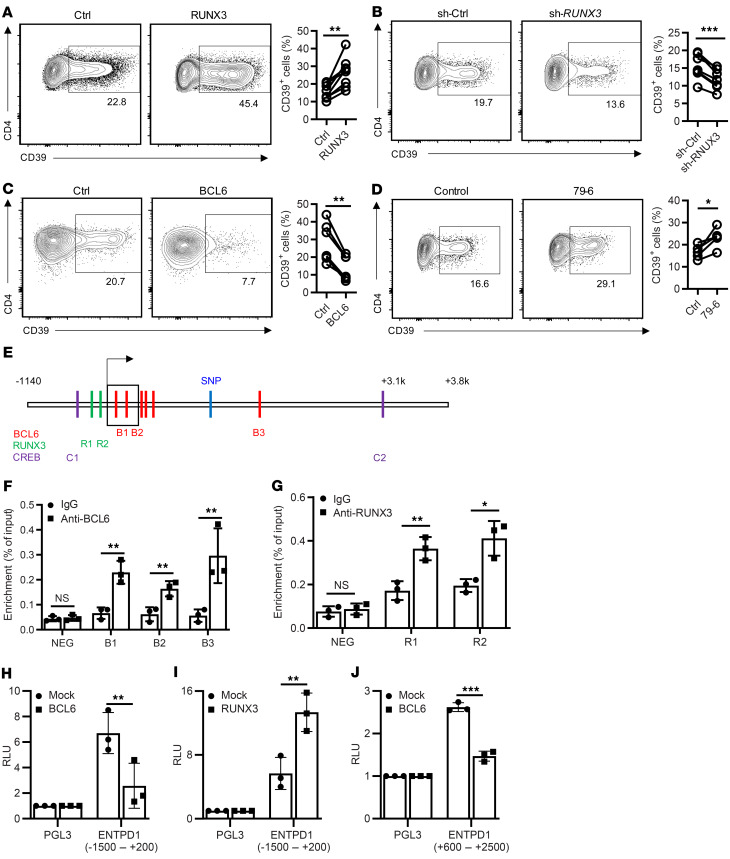
Figure 2. Transcriptional regulation of ENTPD1.
(A–C) Human peripheral T cells were transduced with RUNX3-overexpressing lentivirus (A), RUNX3 shRNA lentivirus (B), or BCL6-overexpressing lentivirus (C). Cells were cultured for 5 days after anti-CD3/anti-CD28 Dynabead activation. CD39 expression in CD4+GFP+ cells was determined by flow cytometry. Results are shown as representative contour plots (left) and summary plots from 6 to 8 experiments (right). (D) Purified human naive CD4+ T cells were activated by anti-CD3/anti-CD28 Dynabeads in the presence or absence of the BCL6 inhibitor 79-6 for 5 days; CD39 expression was determined by flow cytometry. Representative contour plots and summary data from 5 experiments. (E) Predicted BCL6, RUNX3, and p-CREB binding sites adjacent to the ENTPD1 transcription start site. (F and G) Human memory CD4+ cells were activated by anti-CD3/anti-CD28 Dynabeads for 3 days. BCL6 (F) or RUNX3 (G) binding was determined by ChIP-PCR for indicated sites. Results are shown as mean ± SEM of 3 experiments. (H and I) HEK-293T cells were cotransfected with pGL3-basic or pGL3 reporter construct containing the ENTPD1 promoter together with mock or BCL6-overexpressing (H) or RUNX3-overexpressing (I) plasmids. Luciferase activity was determined after 48 hours. Results are shown as mean ± SEM of 3 experiments. (J) Reporter activity in HEK-293T cells cotransfected with pNL3.3-basic or the ENTPD1 enhancer construct together with BCL6 or mock plasmid. Data were compared by 2-tailed paired t test (A–D), 1-way ANOVA with post hoc Tukey’s test (F and G), or unpaired t test (H–J). *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. NS, not significant.