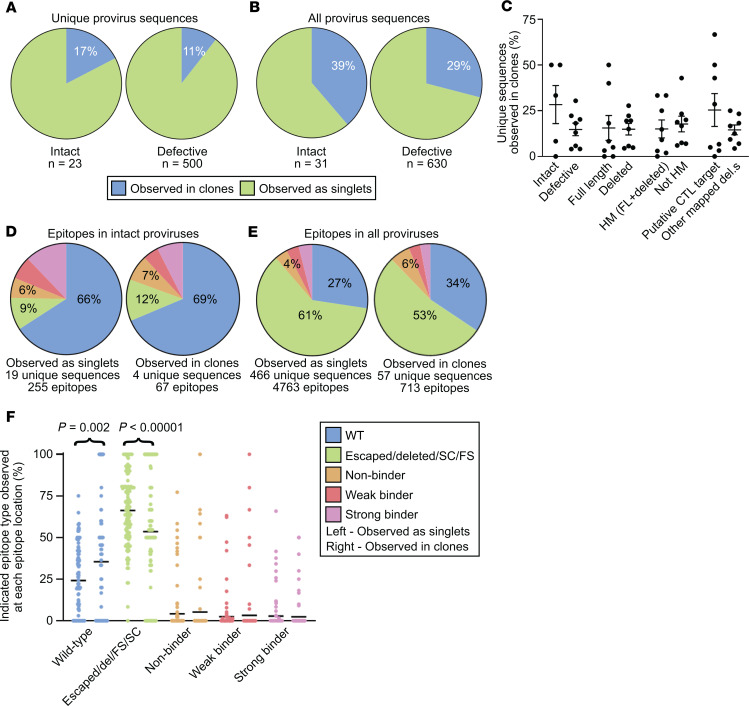
Figure 6. The proportion of intact proviruses observed in large clones is similar to the proportion of defective proviruses observed in large clones.
(A and B) Summary pie charts of all intact (left) and defective (right) proviruses from all participant time points in this study showing proportions observed in clones (blue) versus those observed as singlets (green). In A and C–F, each clone family was collapsed into one unique sequence for counting purposes. In B, clone families were not collapsed. (C) Percentage of proviruses of the indicated type observed in clones relative to total proviruses of the indicated type from each individual, with mean and SEM shown. The putative CTL target grouping includes intact, FL HM, PD, and FS. There was no statistically significant difference in each pair when the nonparametric Kruskal-Wallis test was applied. (D) Summary pie charts of Gag, Pol, and Nef epitope sequences from predicted dominant epitopes from intact proviruses that were observed as singlets (left) or observed in clones (right). (E) Summary pie charts of Gag, Pol, and Nef epitope sequences from predicted dominant epitopes from all proviruses observed as singlets (left) or observed in clones (right). (F) Percentage of indicated epitope type relative to total epitope sequences at each of 125 predicted dominant epitopes from 8 individuals, with mean shown in black. Left: Epitope sequences from proviruses observed as singlets. Right: Epitope sequences from proviruses observed in clones. The nonparametric Wilcoxon’s test was applied and significant P values are shown; see also Supplemental Table 3. D, E, and F share a figure legend. Del, epitope within mapped deletion in provirus; SC or FS, stop codon or frameshift observed between relevant gene’s start codon and epitope; HM, hypermutated; FL, full-length.