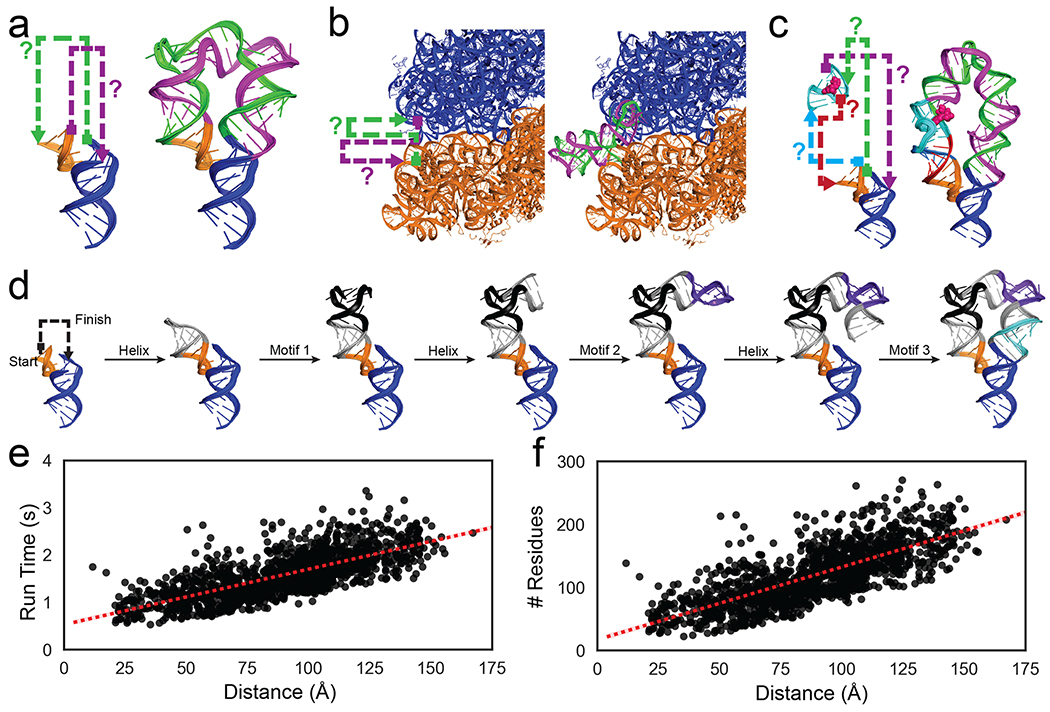
Figure 1: Problems in RNA nanotechnology reduced to RNA motif pathfinding problems and solved by RNAMake.
(a) ‘miniTTRs’ require two strands (green, purple) between tetraloop (orange) and tetraloop-receptor (blue); (b) tethered ribosomes require two strands (green, purple) to link the small subunit (orange) to the large subunit (blue). c) ‘Locking’ a small-molecule binding aptamer (cyan; ATP molecule in pink spheres) by designing four strands (green, purple, teal, magenta) to a peripheral tertiary contact (orange, blue). d) Demonstration of RNAMake design algorithm, which builds an RNA path via the successive addition of motifs and helices from a starting base pair to the ending base pair. e-f) computational efficiency for RNAMake to design connections between each pair of hairpins on the 50S E. coli ribosome. Run time scales linearly with problem size, as measured by (e) translational distance between helical endpoints or (f) number of residues required for segments (see Figure S14 for utilizing higher order junctions).