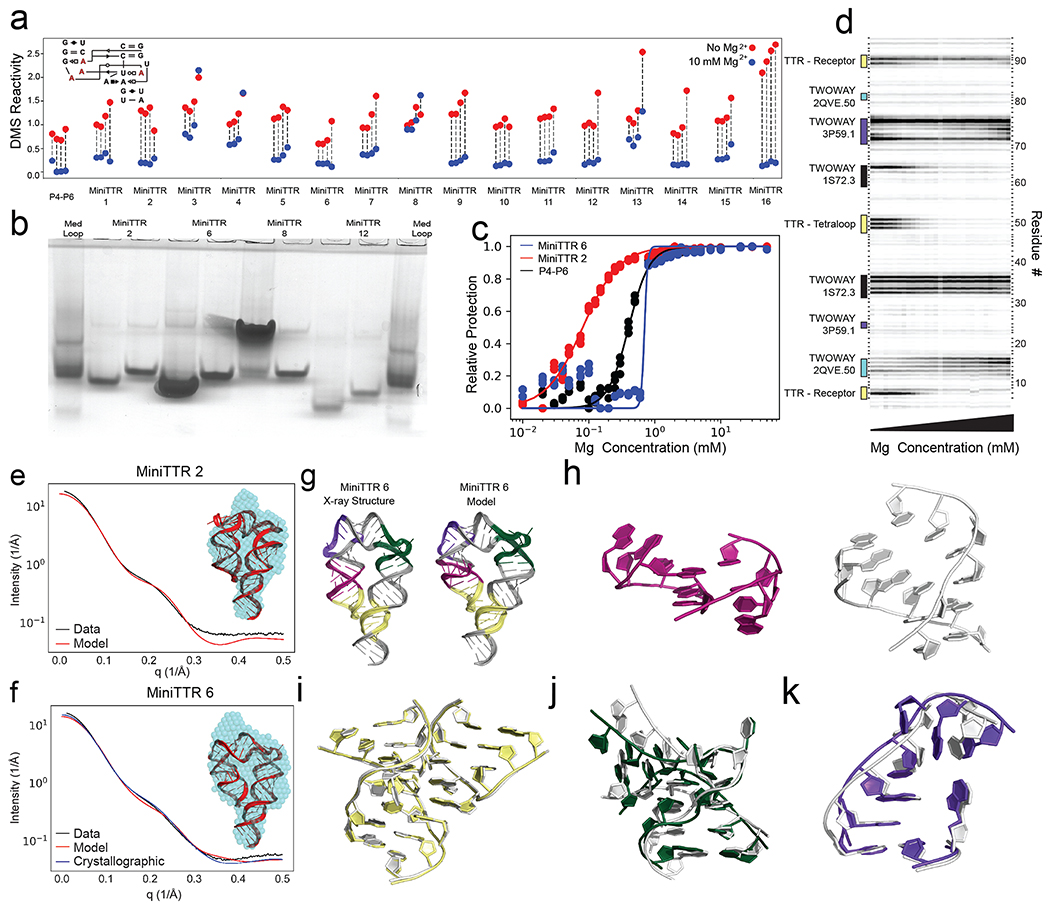
Figure 2: Solving the TTR design problem.
a) Quantification of DMS reactivity in the absence of Mg2+ (red) and with 10 mM Mg2+ (blue) for RNAMake-designed miniTTR constructs and the P4-P6 domain of the Tetrahymena ribozyme as a large natural RNA comparison. Diagram (inset) shows the four adenosines in the tetraloop/tetraloop-receptor (red) that undergo protection upon Mg2+-dependent folding. b) Native gel assays testing whether mutation of GAAA tetraloop (left lane) to UUCG mutant (right lane) disrupts the miniTTR tertiary fold and slows its mobility. ‘MedLoop’ lanes are a control RNA with similar size 58. Figure S3b gives other remaining constructs. c) Quantification of the miniTTR folding stability based on Mg2+ binding curves read out by DMS mapping for miniTTR 6 (blue), miniTTR 2 (red) and P4-P6 (black). d) Raw data from the Mg2+ titration of miniTTR 2, highlighting the change in DMS reactivity in the TTR and the motifs used in the design. e-f) SAXS analysis: Experimental intensity versus scattering amplitude and low-resolution reconstruction derived from experimental scattering profiles (blue beads, inset) overlaid on RNAMake-designed model (cartoon, inset) for (e) miniTTR 2 and (f) miniTTR 6. In (f), SAXS prediction from miniTTR crystal structure is also shown (blue). g-l) X-ray crystal structure of MiniTTR 6 tests accuracy of RNAMake model at atomic resolution: (g) overall RNAMake and X-ray structure, and magnified views of (h) triple mismatch motif from ribosome, (i) tetraloop/tetraloop-receptor, (j) kink-turn motif, and (k) right angle turn. In (h-k), crystal structures are white.