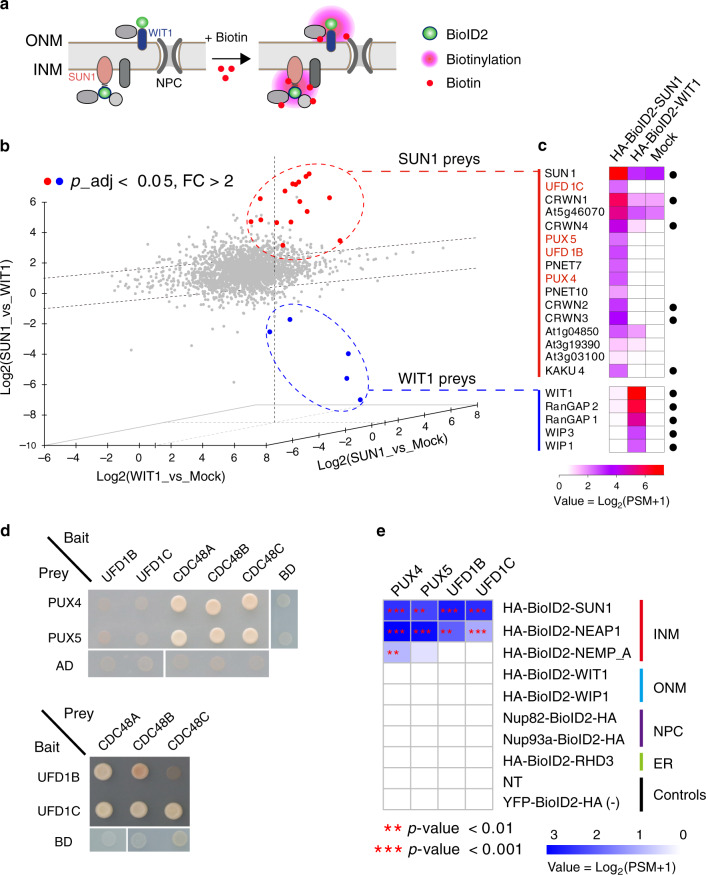
Fig. 1. The CDC48-dependent proteolysis machinery associates with integral INM proteins.
a Identification of proteins that are specifically associated with SUN1 at the INM using proximity labeling. The ONM protein WIT1 serves as a control to exclude proteins associated with both the INM and the ONM. b Pairwise ratiometric analyses of PL-LFQMS data using 35S: HA-BioID2-SUN1 (+biotin), 35S: HA-BioID2-WIT1 (+biotin), and 35S: YFP-BioID2-HA (−biotin) samples. Three biological replicates for each sample were included in the analyses. Axes represent the log values of peptide intensity ratio (FC, fold-change) between two samples. Significantly enriched proteins in one sample were defined by cutoffs FC > 2 and adjusted p-value < 0.05 (linear model F-test) relative to both controls. Proteins significantly enriched in HA-BioID2-SUN1 and HA-BioID2-WIT1 samples are labeled in red and blue dots, respectively. c Heatmap showing transformed and averaged peptide spectrum match (PSM) values of significantly enriched proteins in HA-BioID2-SUN1 and HA-BioID2-WIT1 samples. Known SUN1 and WIT1 interacting proteins are marked by black dots on the right. d Yeast-two-hybrid analyses among PUX4/5, UFD1B/1C, and CDC48A/B/C. Diploid yeasts after mating were grown on QDO medium (SD-Leu/-Trp/-His/-Ade) for 3 days. Similar results have been obtained twice. e Transformed and averaged PSM data of PUX4, PUX5, UFD1B, and UFD1C from PL-LFQMS experiments using BioID2-tagged bait proteins from the inner nuclear membrane (INM), the outer nuclear membrane (ONM), the nuclear pore complex (NPC), and the endoplasmic reticulum (ER). Three biological replicates for each sample were included for analysis. Biotin-treated WT non-transgenic plants (NT) and YFP-BioID2-HA plants without biotin treatment were served as controls. Asterirks (** and ***) represent significant enrichment with p-value < 0.01 and 0.001 (linear model F-test), respectively, compared to both controls. MS data are available through PRIDE (Identifiers PXD015919 and PXD015920) and see Supplementary Data 2 for the dataset list.