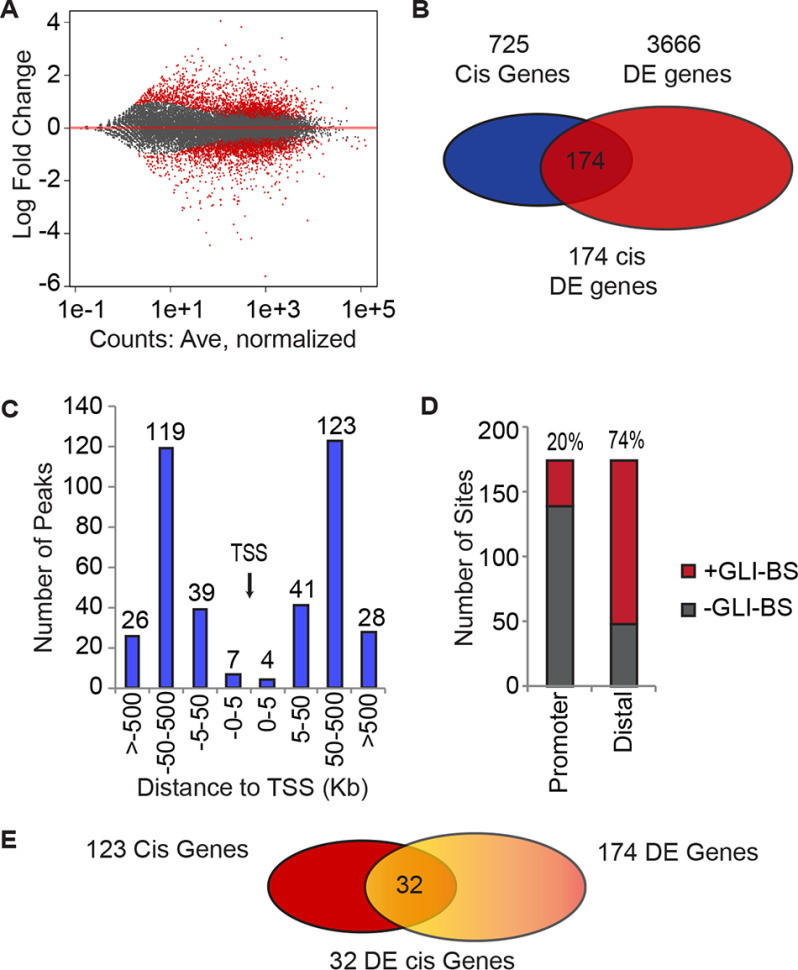
Figure 4.
GLI1-regulated genes have distal putative enhancer regions with GLI-binding site enrichment. A, MA plot showing an approximately equal distribution of up and down-regulated genes following GLI1 knockdown as determined by RNA-seq analysis using DESeq2 at 1.5-fold change (n = 3). B, the differentially expressed genes located cis to GLI1-responsive ATAC peaks were identified. Using GREAT, 725 genes were identified cis to the GLI1 ATAC-seq peaks and then intersected with the 3666 DE GLI1 genes to identify 174 genes. C, the positions of the GLI1-responsive ATAC peaks were mapped relative to the transcription start site for the 174 genes using the GREAT package. D, the frequency of GLI-BS in the promoters and the distal sites of the 174 gene was determined using the HOMER package to identify the GLI-BS by their matrices. E, the differentially expressed genes (32) with distal GLI1- and SMARCA2-responsive ATAC peaks (overlapping) were identified by comparison of the gene lists, 123 genes cis to overlapping GLI1- and SMARCA2-responsive ATAC peaks to the 174 DE genes near a GLI1-responsive ATAC peak.