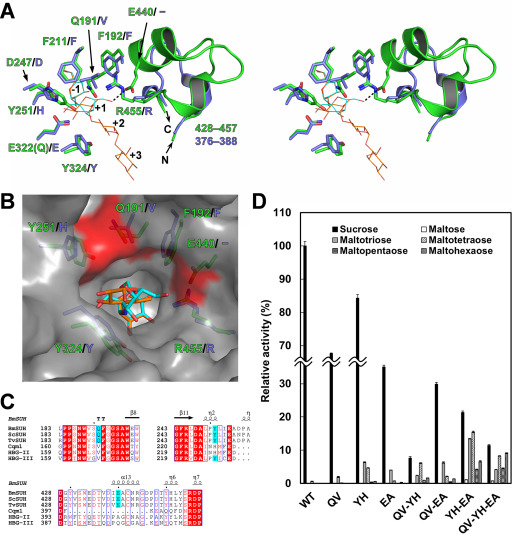
Figure 5.
Amino acid residues important for substrate specificity. A, structural comparison of the active sites of BmSUH (green) and Cqm1 (slate blue) in stereo. The side chains of the amino acid residues around subsite +1 are indicated as sticks, and residues 428–457 of BmSUH and the corresponding region (residues 376–388) of Cqm1 are displayed as ribbon models. Sucrose (cyan) and ACR (orange) derived from E322Q-Suc and BmSUH-ACR are superimposed and indicated as thin stick models. B, molecular surface of the catalytic site of BmSUH. The side chains of the subsite +1 residues (green for BmSUH and slate blue for Cqm1), sucrose (cyan), and an acarviosine moiety of ACR (orange) are indicated. Solvent-accessible areas of Gln191, Tyr251, and Glu440 are highlighted in red. C, sequence alignments of regions around Gln191, Tyr251, and Glu440 and their corresponding regions of GH13_17 sucrose hydrolases and maltases. The conserved residues are highlighted in red; Gln191, Tyr251, and Glu440 of BmSUH and conserved residues of the other GH13_17 enzymes are in cyan. D, hydrolytic activity of BmSUH and its mutants toward sucrose and maltooligosaccharides (from maltose to maltohexaose). Bar charts and error bars, means and S.D., respectively, from triplicate experiments. QV, Q191V; YH, Y251H; EA, E440A; QV-YH, Q191V/Y251H; QV-EA, Q191V/E440A; YH-EA, Y251H/E440A; QV-YH-EA, Q191V/Y251H/E440A.