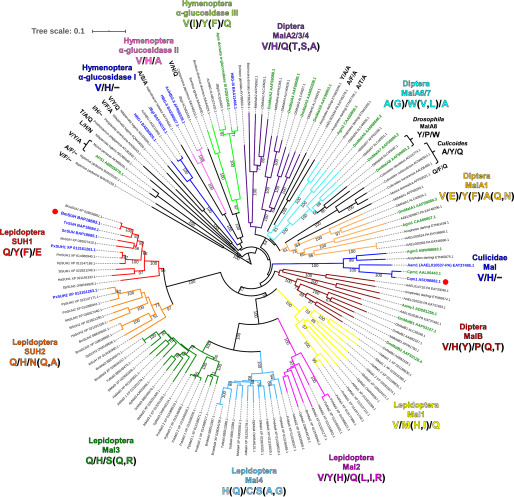
Figure 6.
Phylogenetic tree of GH13_17 proteins and amino acid residues related to substrate specificity. The 142 amino acid sequences were aligned using the MUSCLE program, and the phylogenetic tree constructed by the neighbor-joining method was visualized using the iTOL v5 server. Proteins used were enzymes listed in the CAZy database and their homologs (>40% identity) found using the PSI-BLAST search with BmSUH as a template. Bootstrap values based on 1,000 replicates are shown. Origins, abbreviations, and GenBankTM ID are labeled and summarized in Table S3. GH13_17 proteins are divided into several clades with different colors based on amino acid residues corresponding to subsite +1 (Gln191, Tyr251, and Glu440) in BmSUH. Proteins shown in green and blue are genetically identified and enzymatically characterized, respectively. BmSUH and Cqm1 are marked with red circles.