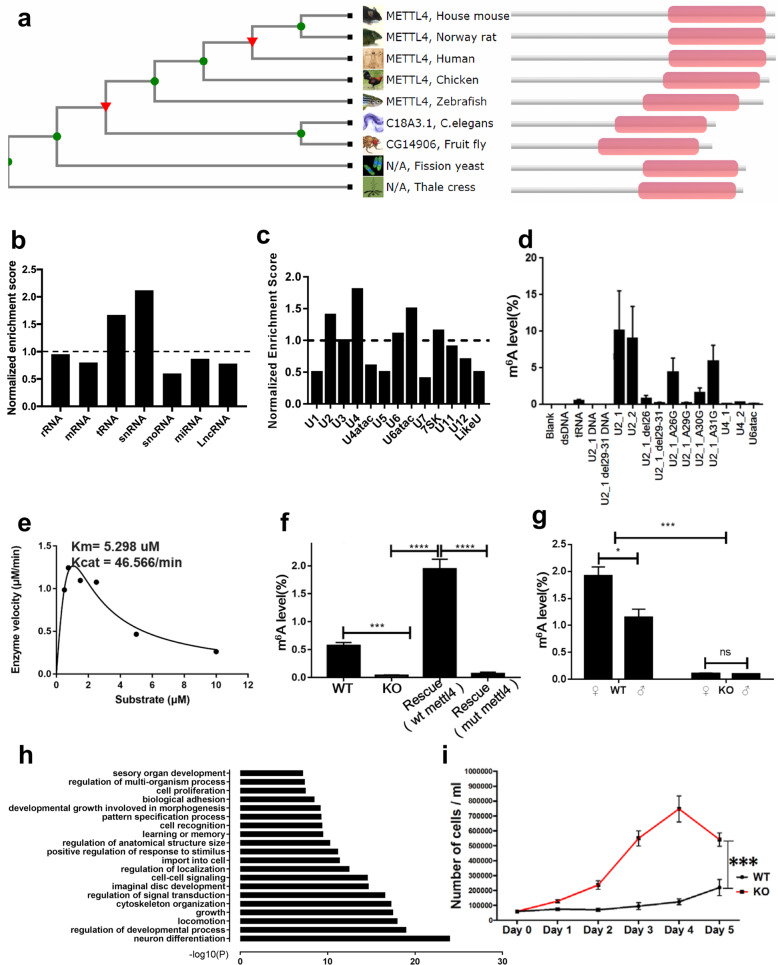
Fig. 1. CG14906 (mettl4) methylates U2 snRNA in Drosophila melanogaster.
a Cladogram of mettl4 in model organisms based on their sequence similarity, the pink rectangle indicates the MT-A70 domain. b Enrichment analysis of eCLIP-seq data for different RNA types. tRNA and snRNA are enriched among all RNA types in general and snRNA is the top enriched RNA molecules targeted by mettl4 in vivo. c Enrichment analysis of eCLIP-seq data for subgroups of snRNA. U2, U4, and U6atac are the top enriched subgroups. d In vitro enzymatic activity is measured by LS-MS/MS using substrates, including U2, U4, U6atac, and U2, with different point mutations and deletions, and tRNA and DNA with U2 sequences. Results show that U2 is the best substrate for fly mettl4 and that adenosine at position 29 in U2 is methylated by mettl4. e Michaelis–Menten kinetics of recombinant mettl4 was determined using U2 probes as substrate by LC-MS/MS analysis. f U2 m6A analysis in WT, KO, and rescued (wt: wild-type mettl4; mut: catalytic dead mutant mettl4, DPPW→NPPW) cells by LC-MS/MS. g In vivo U2 m6A analysis by LC-MS/MS of WT and KO flies. Error bars indicate mean ± SD (n = 3). h Genes with differential alternative splicing were used for the GO analysis. The top 20 enriched biological processes are shown in the bar plot. i Growth curves of WT and mettl4 KO cells in a course of 5 days. Error bars indicate mean ± SD (n = 7). Permutation test was used to determine the significant level of the difference between two groups of growth curves. Statistical significance is determined as: n.s., P > 0.05; *P < 0.05; ***P < 0.001; ****P < 0.0001.